G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition
- PMID: 12235379
- PMCID: PMC137114
- DOI: 10.1093/nar/gkf530
G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition
Abstract
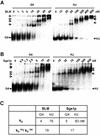
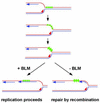
To understand the specific genetic instabilities associated with deficiencies in RecQ family helicases, we have studied the substrate preferences of two closely related members of this family, human BLM and Saccharomyces cerevisiae Sgs1p. Here we show that both BLM and Sgs1p preferentially unwind G4 DNA relative to Holliday junction substrates, and that substrate preference reflects binding affinity and maps to the conserved central helicase domain. We identify the porphyrin N-methyl mesoporphyrin IX (NMM) as a specific inhibitor of G4 DNA unwinding, and show that in the presence of NMM the helicase becomes trapped on the NMM-G4 DNA complex, consuming ATP but unable to unwind or dissociate. These results suggest that BLM and Sgs1p function proactively in replication to remove G4 DNA structures which would otherwise present obstacles to fork progression, rather than by promoting recombination to restart a fork that has stalled.
Figures





References
-
- Ellis N.A., Groden,J., Ye,T.Z., Straughen,J., Lennon,D.J., Ciocci,S., Proytcheva,M. and German,J. (1995) The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell, 83, 655–666. - PubMed
-
- Yu C.E., Oshima,J., Fu,Y.H., Wijsman,E.M., Hisama,F., Alisch,R., Matthews,S., Nakura,J., Miki,T., Ouais,S. et al. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. - PubMed
-
- Kamath-Loeb A.S., Shen,J.C., Loeb,L.A. and Fry,M. (1998) Werner syndrome protein. II. Characterization of the integral 3′→5′ DNA exonuclease. J. Biol. Chem., 273, 34145–34150. - PubMed
-
- Kitao S., Shimamoto,A., Goto,M., Miller,R.W., Smithson,W.A., Lindor,N.M. and Furuichi,Y. (1999) Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson syndrome. Nature Genet., 22, 82–84. - PubMed
-
- Sinclair D.A. and Guarente,L. (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell, 91, 1033–1042. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

