Intrinsic and extrinsic contributions to stochasticity in gene expression
- PMID: 12237400
- PMCID: PMC130539
- DOI: 10.1073/pnas.162041399
Intrinsic and extrinsic contributions to stochasticity in gene expression
Abstract
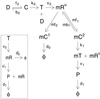
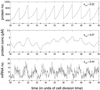
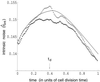
Gene expression is a stochastic, or "noisy," process. This noise comes about in two ways. The inherent stochasticity of biochemical processes such as transcription and translation generates "intrinsic" noise. In addition, fluctuations in the amounts or states of other cellular components lead indirectly to variation in the expression of a particular gene and thus represent "extrinsic" noise. Here, we show how the total variation in the level of expression of a given gene can be decomposed into its intrinsic and extrinsic components. We demonstrate theoretically that simultaneous measurement of two identical genes per cell enables discrimination of these two types of noise. Analytic expressions for intrinsic noise are given for a model that involves all the major steps in transcription and translation. These expressions give the sensitivity to various parameters, quantify the deviation from Poisson statistics, and provide a way of fitting experiment. Transcription dominates the intrinsic noise when the average number of proteins made per mRNA transcript is greater than approximately 2. Below this number, translational effects also become important. Gene replication and cell division, included in the model, cause protein numbers to tend to a limit cycle. We calculate a general form for the extrinsic noise and illustrate it with the particular case of a single fluctuating extrinsic variable-a repressor protein, which acts on the gene of interest. All results are confirmed by stochastic simulation using plausible parameters for Escherichia coli.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

