A signal transduction protein cues proteolytic events critical to Caulobacter cell cycle progression
- PMID: 12237413
- PMCID: PMC130603
- DOI: 10.1073/pnas.202495099
A signal transduction protein cues proteolytic events critical to Caulobacter cell cycle progression
Abstract
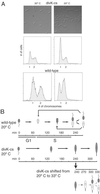
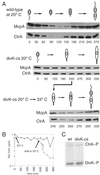
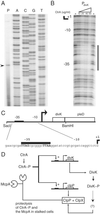
Temporally controlled proteolysis of the essential response regulator, CtrA, is critical for cell cycle progression in Caulobacter crescentus. CtrA binds to and silences the origin of replication in swarmer cells. The initiation of replication depends on the proteolysis of CtrA. We present evidence that DivK, an essential single-domain response regulator, contributes to the control of the G(1)-S transition by signaling the temporally controlled proteolysis of CtrA. In a divK-cs mutant at the restrictive temperature, the initiation of DNA replication is blocked because of the retention of CtrA. A shift of cells from restrictive to permissive temperature results in rapid degradation of CtrA, initiation of DNA replication, and the resumption of cell cycle progression, including the ordered expression of genes involved in chromosome replication and polar organelle biogenesis. CtrA binds to and regulates the promoters of two genes critical to its temporally controlled proteolysis, divK and clpP, providing a transcriptional feedback loop for the control of cell cycle progression.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

