The origins of asymmetry in the folding transition states of protein L and protein G
- PMID: 12237457
- PMCID: PMC2373711
- DOI: 10.1110/ps.0205402
The origins of asymmetry in the folding transition states of protein L and protein G
Abstract
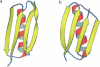
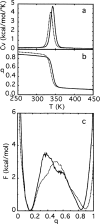
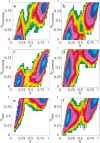
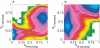
Topology has been shown to be an important determinant of many features of protein folding; however, the delineation of sequence effects on folding remains obscure. Furthermore, differentiation between the two influences proves difficult due to their intimate relationship. To investigate the effect of sequence in the absence of significant topological differences, we examined the folding mechanisms of segment B1 peptostreptococcal protein L and segment B1 of streptococcal protein G. These proteins share the same highly symmetrical topology. Despite this symmetry, neither protein folds through a symmetrical transition state. We analyzed the origins of this difference using theoretical models. We found that the strength of the interactions present in the N-terminal hairpin of protein L causes this hairpin to form ahead of the C-terminal hairpin. The difference in chain entropy associated with the formation of the hairpins of protein G proves sufficient to beget initiation of folding at the shorter C-terminal hairpin. Our findings suggest that the mechanism of folding may be understood by examination of the free energy associated with the formation of partially folded microstates.
Figures
Similar articles
-
Critical role of beta-hairpin formation in protein G folding.Nat Struct Biol. 2000 Aug;7(8):669-73. doi: 10.1038/77971. Nat Struct Biol. 2000. PMID: 10932252
-
A breakdown of symmetry in the folding transition state of protein L.J Mol Biol. 2000 May 19;298(5):971-84. doi: 10.1006/jmbi.2000.3701. J Mol Biol. 2000. PMID: 10801362
-
Interplay among tertiary contacts, secondary structure formation and side-chain packing in the protein folding mechanism: all-atom representation study of protein L.J Mol Biol. 2003 Feb 21;326(3):933-54. doi: 10.1016/s0022-2836(02)01379-7. J Mol Biol. 2003. PMID: 12581651
-
Protein folding and the organization of the protein topology universe.Trends Biochem Sci. 2005 Jan;30(1):13-9. doi: 10.1016/j.tibs.2004.11.008. Trends Biochem Sci. 2005. PMID: 15653321 Review.
-
Circuit Topology Analysis of Polymer Folding Reactions.ACS Cent Sci. 2020 Jun 24;6(6):839-847. doi: 10.1021/acscentsci.0c00308. Epub 2020 May 12. ACS Cent Sci. 2020. PMID: 32607431 Free PMC article. Review.
Cited by
-
Coarse-grained models reveal functional dynamics--II. Molecular dynamics simulation at the coarse-grained level--theories and biological applications.Bioinform Biol Insights. 2008 Mar 5;2:171-85. doi: 10.4137/bbi.s459. Bioinform Biol Insights. 2008. PMID: 19812774 Free PMC article.
-
Capturing the essence of folding and functions of biomolecules using coarse-grained models.Nat Commun. 2011 Sep 27;2:487. doi: 10.1038/ncomms1481. Nat Commun. 2011. PMID: 21952221 Review.
-
Building a macro-mixing dual-basin Gō model using the Multistate Bennett Acceptance Ratio.Biophys Physicobiol. 2019 Nov 29;16:310-321. doi: 10.2142/biophysico.16.0_310. eCollection 2019. Biophys Physicobiol. 2019. PMID: 31984186 Free PMC article.
-
Visualizing chaperone-assisted protein folding.Nat Struct Mol Biol. 2016 Jul;23(7):691-7. doi: 10.1038/nsmb.3237. Epub 2016 May 30. Nat Struct Mol Biol. 2016. PMID: 27239796 Free PMC article.
-
Multiple-Basin Go̅-Martini for Investigating Conformational Transitions and Environmental Interactions of Proteins.J Chem Theory Comput. 2025 May 27;21(10):5304-5321. doi: 10.1021/acs.jctc.5c00256. Epub 2025 May 13. J Chem Theory Comput. 2025. PMID: 40359486 Free PMC article.
References
-
- Alexander, P., Orban, J., and Bryan, P. 1992. Kinetic analysis of folding and unfolding the 56 amino acid IgG-binding domain of streptococcal protein G. Biochemistry 31 7243–7248. - PubMed
-
- Bernstein, F., Koetzle, T., Williams, G., Meyer, E., Brice, M., Rodgers, J., Kennard, O., Shimanouchi, T., and Tasumi, M. 1977. The Protein Data Bank: A computer-based archival file for macromolecular structures. J. Mol. Biol. 112 535–542. - PubMed
-
- Bilsel, O. and Matthews, C.R. 2000. Barriers in protein folding reactions. Adv. Prot. Chem. 53 153–207. - PubMed
-
- Blanco, F.J. and Serrano, L. 1995. Folding of protein G B1 domain studied by the conformational characterization of fragments comprising its secondary structure elements. Eur. J. Biochem. 230 634–649. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

