Quantitation of human immunodeficiency virus type 1 DNA forms with the second template switch in peripheral blood cells predicts disease progression independently of plasma RNA load
- PMID: 12239284
- PMCID: PMC136544
- DOI: 10.1128/jvi.76.20.10099-10108.2002
Quantitation of human immunodeficiency virus type 1 DNA forms with the second template switch in peripheral blood cells predicts disease progression independently of plasma RNA load
Abstract
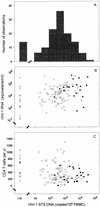
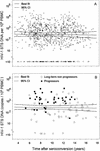
There are several forms of human immunodeficiency virus type 1 (HIV-1) DNA in peripheral blood T cells and lymph nodes in untreated HIV-1-infected individuals and in patients whose plasma HIV-1 RNA levels are suppressed by long-term combination antiretroviral therapy. However, it remains to be established whether the concentration of HIV-1 DNA in cells predicts the clinical outcome of HIV-1 infection. In this report, we measured the concentration of HIV-1 DNA forms which has undergone the second template switch (STS DNA) and 2-long-terminal-repeat DNA circles in peripheral blood mononuclear cell (PBMC) samples. To do this, we used molecular-beacon-based real-time PCR assays and studied 130 patients with hemophilia in the Multicenter Hemophilia Cohort Study. We assessed the influence of baseline HIV-1 STS DNA levels on the progression of HIV-1 disease in the absence of combination antiretroviral therapy by Kaplan-Meier and Cox regression analysis. Among the patients who progressed to AIDS, the median levels (interquartile ranges) of STS HIV-1 DNA in PBMC were significantly higher than those of patients who remained AIDS free during the 16 years of follow-up (1,017 [235 to 6,059] and 286 [31 to 732] copies per 10(6) PBMC, respectively; P < 0.0001). Rates of progression to death and development of AIDS varied significantly (log rank P < 0.001) by quartile distribution of HIV-1 STS DNA levels. After adjustment for age at seroconversion, baseline CD4(+) T-cell counts, plasma viral load, and T-cell-receptor excision circles, the relative hazards (RH) of death and AIDS were significantly increased with higher HIV-1 STS DNA levels (adjusted RH, 1.84 [95% confidence interval (CI), 1.30 to 2.59] and 2.62 [95% CI, 1.75 to 3.93] per 10-fold increase per 10(6) PBMC, respectively). HIV-1 STS DNA levels in each individual remained steady in longitudinal PBMC samples during 16 years of follow-up. Our findings show that the concentration of HIV-1 STS DNA in PBMC complements the HIV-1 RNA load in plasma in predicting the clinical outcome of HIV-1 disease. This parameter may have important implications for understanding the virological response to combination antiretroviral therapy.
Figures


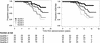

References
-
- Aoki, S., R. Yarchoan, R. V. Thomas, J. M. Pluda, K. Marczyk, S. Broder, and H. Mitsuya. 1990. Quantitative analysis of HIV-1 proviral DNA in peripheral blood mononuclear cells from patients with AIDS or ARC: decrease of proviral DNA content following treatment with 2′,3′-dideoxyinosine (ddI). AIDS Res. Hum. Retrovir. 6:1331-1339. - PubMed
-
- Bieniasz, P. D., K. Ariyoshi, M. A. Bourelly, S. Bloor, R. B. Foxall, E. C. Harwood, and J. N. Weber. 1993. Variable relationship between proviral DNA load and infectious virus titre in the peripheral blood mononuclear cells of HIV-1-infected individuals. AIDS 7:803-806. - PubMed
-
- Bruisten, S. M., P. Reiss, A. E. Loeliger, P. van Swieten, R. Schuurman, C. A. Boucher, G. J. Weverling, and J. G. Huisman. 1998. Cellular proviral HIV type 1 DNA load persists after long-term RT-inhibitor therapy in HIV type 1 infected persons. AIDS Res. Hum. Retrovir. 14:1053-1058. - PubMed
-
- Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

