Evidence against a simple tethering model for enhancement of herpes simplex virus DNA polymerase processivity by accessory protein UL42
- PMID: 12239303
- PMCID: PMC136589
- DOI: 10.1128/jvi.76.20.10270-10281.2002
Evidence against a simple tethering model for enhancement of herpes simplex virus DNA polymerase processivity by accessory protein UL42
Abstract
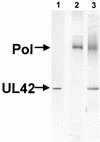
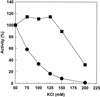
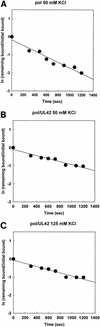
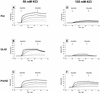
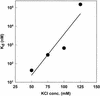
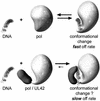
The DNA polymerase holoenzyme of herpes simplex virus type 1 (HSV-1) is a stable heterodimer consisting of a catalytic subunit (Pol) and a processivity factor (UL42). HSV-1 UL42 differs from most DNA polymerase processivity factors in possessing an inherent ability to bind to double-stranded DNA. It has been proposed that UL42 increases the processivity of Pol by directly tethering it to the primer and template (P/T). To test this hypothesis, we took advantage of the different sensitivities of Pol and Pol/UL42 activities to ionic strength. Although the activity of Pol is inhibited by salt concentrations in excess of 50 mM KCl, the activity of the holoenzyme is relatively refractory to changes in ionic strength from 50 to 125 mM KCl. We used nitrocellulose filter-binding assays and real-time biosensor technology to measure binding affinities and dissociation rate constants of the individual subunits and holoenzyme for a short model P/T as a function of the ionic strength of the buffer. We found that as observed for activity, the binding affinity and dissociation rate constant of the Pol/UL42 holoenzyme for P/T were not altered substantially in high- versus low-ionic-strength buffer. In 50 mM KCl, the apparent affinity with which UL42 bound the P/T did not differ by more than twofold compared to that observed for Pol or Pol/UL42 in the same low-ionic-strength buffer. However, increasing the ionic strength dramatically decreased the affinity of UL42 for P/T, such that it was reduced more than 3 orders of magnitude from that of Pol/UL42 in 125 mM KCl. Real-time binding kinetics revealed that much of the reduced affinity could be attributable to an extremely rapid dissociation of UL42 from the P/T in high-ionic-strength buffer. The resistance of the activity, binding affinity, and stability of the holoenzyme for the model P/T to increases in ionic strength, despite the low apparent affinity and poor stability with which UL42 binds the model P/T in high concentrations of salt, suggests that UL42 does not simply tether the Pol to DNA. Instead, it is likely that conformational alterations induced by interaction of UL42 with Pol allow for high-affinity and high-stability binding of the holoenzyme to the P/T even under high-ionic-strength conditions.
Figures
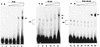

Similar articles
-
Interaction of herpes simplex virus type 1 DNA polymerase and the UL42 accessory protein with a model primer template.J Virol. 1994 Aug;68(8):4937-45. doi: 10.1128/JVI.68.8.4937-4945.1994. J Virol. 1994. PMID: 8035492 Free PMC article.
-
Effects of substitutions of arginine residues on the basic surface of herpes simplex virus UL42 support a role for DNA binding in processive DNA synthesis.J Virol. 2005 Sep;79(18):12025-34. doi: 10.1128/JVI.79.18.12025-12034.2005. J Virol. 2005. PMID: 16140778 Free PMC article.
-
Herpes simplex virus processivity factor UL42 imparts increased DNA-binding specificity to the viral DNA polymerase and decreased dissociation from primer-template without reducing the elongation rate.J Virol. 1999 Jan;73(1):55-66. doi: 10.1128/JVI.73.1.55-66.1999. J Virol. 1999. PMID: 9847307 Free PMC article.
-
DNA and protein interactions of the small subunit of herpes simplex virus type 1 DNA polymerase.Virology. 1999 Jan 5;253(1):55-64. doi: 10.1006/viro.1998.9491. Virology. 1999. PMID: 9887318
-
Herpesvirus DNA polymerase processivity factors: Not just for DNA synthesis.Virus Res. 2021 Jun;298:198394. doi: 10.1016/j.virusres.2021.198394. Epub 2021 Mar 26. Virus Res. 2021. PMID: 33775751 Review.
Cited by
-
Herpes simplex virus 1 DNA polymerase processivity factor UL42 inhibits TNF-α-induced NF-κB activation by interacting with p65/RelA and p50/NF-κB1.Med Microbiol Immunol. 2013 Aug;202(4):313-25. doi: 10.1007/s00430-013-0295-0. Epub 2013 May 1. Med Microbiol Immunol. 2013. PMID: 23636254
-
3' to 5' exonuclease activity of herpes simplex virus type 1 DNA polymerase modulates its strand displacement activity.J Virol. 2003 Sep;77(18):10147-53. doi: 10.1128/jvi.77.18.10147-10153.2003. J Virol. 2003. PMID: 12941927 Free PMC article.
-
White spot syndrome virus Orf514 encodes a bona fide DNA polymerase.Molecules. 2011 Jan 12;16(1):532-42. doi: 10.3390/molecules16010532. Molecules. 2011. PMID: 21228759 Free PMC article.
-
Processing of lagging-strand intermediates in vitro by herpes simplex virus type 1 DNA polymerase.J Virol. 2010 Aug;84(15):7459-72. doi: 10.1128/JVI.01875-09. Epub 2010 May 5. J Virol. 2010. PMID: 20444887 Free PMC article.
-
Changes in subcellular localization reveal interactions between human cytomegalovirus terminase subunits.Virol J. 2012 Dec 21;9:315. doi: 10.1186/1743-422X-9-315. Virol J. 2012. PMID: 23259714 Free PMC article.
References
-
- Bondeson, K., A. Frostell-Karlsson, L. Fagerstam, and G. Magnusson. 1993. Lactose repressor-operator DNA interactions: kinetic analysis by a surface plasmon resonance biosensor. Anal. Biochem. 214:245-251. - PubMed
-
- Bridges, K. G., Q.-X. Hua, M. R. Brigham-Burke, J. D. Martin, P. Hensley, C. E. Dahl, P. Digard, M. A. Weiss, and D. M. Coen. 2000. Secondary structure and structure-activity relationships of peptides corresponding to the subunit interface of herpes simplex virus DNA polymerase. J. Biol. Chem. 275:472-478. - PubMed
-
- Burgers, P. M. J., and B. L. Yoder. 1993. ATP-independent loading of the proliferating cell nuclear antigen requires DNA ends. J. Biol. Chem. 268:19923-19926. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

