Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins
- PMID: 12242332
- PMCID: PMC130532
- DOI: 10.1073/pnas.162488899
Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins
Abstract
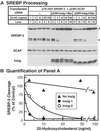
This paper describes insig-2, a second protein of the endoplasmic reticulum that blocks the processing of sterol regulatory element-binding proteins (SREBPs) by binding to SCAP (SREBP cleavage-activating protein) in a sterol-regulated fashion, thus preventing it from escorting SREBPs to the Golgi. By blocking this movement, insig-2, like the previously described insig-1, prevents the proteolytic processing of SREBPs by Golgi enzymes, thereby blocking cholesterol synthesis. The sequences of human insig-1 and -2 are 59% identical. Both proteins are predicted to contain six transmembrane helices. The proteins differ functionally in two respects: (i) production of insig-1, but not insig-2, in cultured mammalian cells requires nuclear SREBPs; and (ii) at high levels of expression, insig-1, but not insig-2, can block SCAP movement in the absence of exogenous sterols. The combined actions of insig-1 and -2 permit feedback regulation of cholesterol synthesis over a wide range of sterol concentrations.
Figures






References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

