Bacterial discrimination by means of a universal array approach mediated by LDR (ligase detection reaction)
- PMID: 12243651
- PMCID: PMC128835
- DOI: 10.1186/1471-2180-2-27
Bacterial discrimination by means of a universal array approach mediated by LDR (ligase detection reaction)
Abstract
Background: PCR amplification of bacterial 16S rRNA genes provides the most comprehensive and flexible means of sampling bacterial communities. Sequence analysis of these cloned fragments can provide a qualitative and quantitative insight of the microbial population under scrutiny although this approach is not suited to large-scale screenings. Other methods, such as denaturing gradient gel electrophoresis, heteroduplex or terminal restriction fragment analysis are rapid and therefore amenable to field-scale experiments. A very recent addition to these analytical tools is represented by microarray technology.
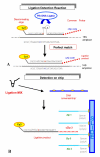
Results: Here we present our results using a Universal DNA Microarray approach as an analytical tool for bacterial discrimination. The proposed procedure is based on the properties of the DNA ligation reaction and requires the design of two probes specific for each target sequence. One oligo carries a fluorescent label and the other a unique sequence (cZipCode or complementary ZipCode) which identifies a ligation product. Ligated fragments, obtained in presence of a proper template (a PCR amplified fragment of the 16s rRNA gene) contain either the fluorescent label or the unique sequence and therefore are addressed to the location on the microarray where the ZipCode sequence has been spotted. Such an array is therefore "Universal" being unrelated to a specific molecular analysis. Here we present the design of probes specific for some groups of bacteria and their application to bacterial diagnostics.
Conclusions: The combined use of selective probes, ligation reaction and the Universal Array approach yielded an analytical procedure with a good power of discrimination among bacteria.
Figures





References
-
- Espejo RT, Feijoo CG, Romero J, Vasquez M. PAGE analysis of the heteroduplexes formed between PCR-amplified 16S rRNA genes: estimation of sequence similarity and rDNA complexity. Microbiology. 1998;144:1611–1617. - PubMed
-
- Delwart EL, Shpaer EG, Louwagie J, McCutcham FE, Grez M, Rubsamen-Waigmann H, Mullins JI. Genetic relationships determined by a DNA heteroduplex mobility assay analysis of HIV-1 env genes. Science. 1993;262:1257–1261. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

