Genes encoding specific nickel transport systems flank the chromosomal urease locus of pathogenic yersiniae
- PMID: 12270829
- PMCID: PMC139606
- DOI: 10.1128/JB.184.20.5706-5713.2002
Genes encoding specific nickel transport systems flank the chromosomal urease locus of pathogenic yersiniae
Abstract
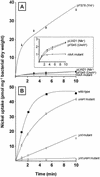
The transition metal nickel is an essential cofactor for a number of bacterial enzymes, one of which is urease. Prior to its incorporation into metalloenzyme active sites, nickel must be imported into the cell. Here, we report identification of two loci corresponding to nickel-specific transport systems in the gram-negative, ureolytic bacterium Yersinia pseudotuberculosis. The loci are located on each side of the chromosomal urease gene cluster ureABCEFGD and have the same orientation as the latter. The yntABCDE locus upstream of the ure genes encodes five predicted products with sequence homology to ATP-binding cassette nickel permeases present in several gram-negative bacteria. The ureH gene, located downstream of ure, encodes a single-component carrier which displays homology to polypeptides of the nickel-cobalt transporter family. Transporters with homology to these two classes are also present (again in proximity to the urease locus) in the other two pathogenic yersiniae, Y. pestis and Y. enterocolitica. An Escherichia coli nikA insertion mutant recovered nickel uptake ability following heterologous complementation with either the ynt or the ureH plasmid-borne gene of Y. pseudotuberculosis, demonstrating that each carrier is necessary and sufficient for nickel transport. Deletion of ynt in Y. pseudotuberculosis almost completely abolished bacterial urease activity, whereas deletion of ureH had no effect. Nevertheless, rates of nickel transport were significantly altered in both ynt and ureH mutants. Furthermore, the ynt ureH double mutant was totally devoid of nickel uptake ability, thus indicating that Ynt and UreH constitute the only routes for nickel entry. Both Ynt and UreH show selectivity for Ni(2+) ions. This is the first reported identification of genes coding for both kinds of nickel-specific permeases situated adjacent to the urease gene cluster in the genome of a microorganism.
Figures



References
-
- Chivers, P. T., and R. T. Sauer. 2000. Regulation of high affinity nickel uptake in bacteria. Ni2+-dependent interaction of NikR with wild-type and mutant operator sites. J. Biol. Chem. 275:19735-19741. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

