Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures
- PMID: 12270832
- PMCID: PMC139613
- DOI: 10.1128/JB.184.20.5733-5745.2002
Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures
Abstract
This work assesses relationships for 30 complete prokaryotic genomes between the presence of the Shine-Dalgarno (SD) sequence and other gene features, including expression levels, type of start codon, and distance between successive genes. A significant positive correlation of the presence of an SD sequence and the predicted expression level of a gene based on codon usage biases was ascertained, such that predicted highly expressed genes are more likely to possess a strong SD sequence than average genes. Genes with AUG start codons are more likely than genes with other start codons, GUG or UUG, to possess an SD sequence. Genes in close proximity to upstream genes on the same coding strand in most genomes are significantly higher in SD presence. In light of these results, we discuss the role of the SD sequence in translation initiation and its relationship with predicted gene expression levels and with operon structure in both bacterial and archaeal genomes.
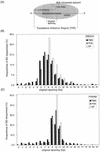
Figures



References
-
- Antelmann, H., J. Bernhardt, R. Schmid, H. Mach, U. Volker, and M. Hecker. 1997. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis 18:1451-1463. - PubMed
-
- Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905-920. - PubMed
-
- Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. - PubMed
-
- Brodersen, D. E., W. M. Clemons, Jr., A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143-1154. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

