Minimum molecular architectures for transcription and replication of the influenza virus
- PMID: 12271117
- PMCID: PMC130604
- DOI: 10.1073/pnas.152456799
Minimum molecular architectures for transcription and replication of the influenza virus
Abstract
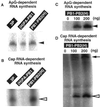
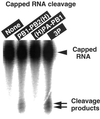
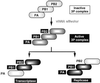
The RNA-dependent RNA polymerase of influenza virus is composed of three viral P proteins (PB1, PB2, and PA) and involved in both transcription and replication of the RNA genome. The PB1 subunit plays a key role in both the assembly of three P protein subunits and the catalytic function of RNA polymerization. We have established a simultaneous expression system of three P proteins in various combinations using recombinant baculoviruses, and isolated the PA-PB1-PB2 ternary (3P) complex and two kinds of the binary (2P) complex, PA-PB1 and PB1-PB2. The affinity-purified 3P complex showed all of the catalytic properties characteristic of the transcriptase, including capped RNA-binding, capped RNA cleavage, model viral RNA binding, model viral RNA-directed RNA synthesis, and polyadenylation of newly synthesized RNA. The PB1-PB2 binary complex showed essentially the same catalytic properties as does the 3P complex, whereas the PA-PB1 complex catalyzed de novo initiation of RNA synthesis in the absence of primers. Taken together we propose that the catalytic specificity of PB1 subunit is modulated to the transcriptase by binding PB2 or the replicase by interaction with PA.
Figures

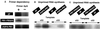

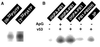
References
-
- Palese P, Kingsbury D W, editors. Genetics of Influenza Viruses. Vienna: Springer; 1983.
-
- Krug R M, Alonso-Caplen F V, Julkenun I, Katze M G. In: The Influenza Viruses. Krug R M, editor. New York: Plenum; 1989. pp. 89–152.
-
- Ishihama A. Biochimie. 1996;78:1097–1102. - PubMed
-
- Honda A, Ishihama A. Biol Chem. 1997;378:483–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

