Distinct antigenic features of linear epitopes at the N-terminus and C-terminus of 65 kDa glutamic acid decarboxylase (GAD65): implications for autoantigen modification during pathogenesis
- PMID: 12296864
- PMCID: PMC1906485
- DOI: 10.1046/j.1365-2249.2002.01960.x
Distinct antigenic features of linear epitopes at the N-terminus and C-terminus of 65 kDa glutamic acid decarboxylase (GAD65): implications for autoantigen modification during pathogenesis
Abstract
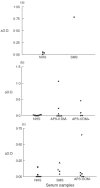
Autoantibodies to 65 kDa glutamic acid decarboxylase (GAD65) are produced in many patients with autoimmune polyendocrine syndrome type II (APS-II) or stiff-man syndrome (SMS) and are heterogeneous in their epitope specificities, recognizing both conformational and linear determinants. Major linear epitopes of GAD, which are recognized by autoantibodies in a minority of these patients, occur in the N-terminal and C-terminal regions. We have investigated antibody recognition of the N- and C-termini of GAD65 in relation to their structural features as an approach to understanding what modifications to the native GAD structure may occur that facilitate the generation of antibodies specific to linear epitopes in these regions during the autoimmune pathogenesis. A monoclonal antibody specific to the N-terminus of GAD65 bound both native and denatured GAD in ELISA, whereas monoclonal and polyclonal antibodies specific to the C-terminus of GAD bound only denatured GAD. These antibodies were epitope mapped using random peptide phage-display libraries and the epitopes related to a previously proposed structural model of GAD65. This has led us to propose that the alpha-helical secondary structure of the C-terminus of GAD65 must be denatured to generate linear epitopes. In contrast, the N-terminus is both surface exposed and linear in the native structure, but may be masked by membrane interactions, which must be broken to facilitate recognition by B cells.
Figures



Similar articles
-
Conformational epitopes on the diabetes autoantigen GAD65 identified by peptide phage display and molecular modeling.J Immunol. 2000 Oct 1;165(7):3830-8. doi: 10.4049/jimmunol.165.7.3830. J Immunol. 2000. PMID: 11034389
-
Comparative analysis of epitope recognition of glutamic acid decarboxylase (GAD) by autoantibodies from different autoimmune disorders.Clin Exp Immunol. 1999 Dec;118(3):349-56. doi: 10.1046/j.1365-2249.1999.01030.x. Clin Exp Immunol. 1999. PMID: 10594551 Free PMC article.
-
Higher autoantibody levels and recognition of a linear NH2-terminal epitope in the autoantigen GAD65, distinguish stiff-man syndrome from insulin-dependent diabetes mellitus.J Exp Med. 1994 Aug 1;180(2):595-606. doi: 10.1084/jem.180.2.595. J Exp Med. 1994. PMID: 7519242 Free PMC article.
-
Structural biology of the GAD autoantigen.Autoimmun Rev. 2010 Jan;9(3):148-52. doi: 10.1016/j.autrev.2009.05.003. Epub 2009 May 22. Autoimmun Rev. 2010. PMID: 19465164 Review.
-
Stiff-person syndrome (SPS) and anti-GAD-related CNS degenerations: protean additions to the autoimmune central neuropathies.J Autoimmun. 2011 Sep;37(2):79-87. doi: 10.1016/j.jaut.2011.05.005. Epub 2011 Jun 16. J Autoimmun. 2011. PMID: 21680149 Review.
Cited by
-
Post-translational protein modifications in type 1 diabetes: a role for the repair enzyme protein-L-isoaspartate (D-aspartate) O-methyltransferase?Diabetologia. 2007 Mar;50(3):676-81. doi: 10.1007/s00125-006-0556-1. Epub 2007 Jan 10. Diabetologia. 2007. PMID: 17216280
-
Identification of new autoantigens by protein array indicates a role for IL4 neutralization in autoimmune hepatitis.Mol Cell Proteomics. 2012 Dec;11(12):1885-97. doi: 10.1074/mcp.M112.018713. Epub 2012 Sep 20. Mol Cell Proteomics. 2012. PMID: 22997428 Free PMC article.
-
Rat β₃-adrenoceptor protein expression: antibody validation and distribution in rat gastrointestinal and urogenital tissues.Naunyn Schmiedebergs Arch Pharmacol. 2014 Nov;387(11):1117-27. doi: 10.1007/s00210-014-1039-4. Epub 2014 Aug 30. Naunyn Schmiedebergs Arch Pharmacol. 2014. PMID: 25172522
-
High-Density Peptide Microarray Analysis of IgG Autoantibody Reactivities in Serum and Cerebrospinal Fluid of Multiple Sclerosis Patients.Mol Cell Proteomics. 2016 Apr;15(4):1360-80. doi: 10.1074/mcp.M115.051664. Epub 2016 Feb 1. Mol Cell Proteomics. 2016. PMID: 26831522 Free PMC article.
-
GAD antibody-spectrum disorders: progress in clinical phenotypes, immunopathogenesis and therapeutic interventions.Ther Adv Neurol Disord. 2021 Mar 30;14:17562864211003486. doi: 10.1177/17562864211003486. eCollection 2021. Ther Adv Neurol Disord. 2021. PMID: 33854562 Free PMC article. Review.
References
-
- Leslie RDG, Atkinson MA, Notkins AL. Autoantigens IA-2 and GAD in type 1 (insulin-dependent) diabetes. Diabetologia. 1999;42:3–14. - PubMed
-
- Kanaani J, Lissin D, Kash SF, Baekkeskov S. The hydrophilic isoform of glutamate decarboxylase, GAD67, is targeted to membranes and nerve terminals independent of dimerization with the hydrophilic membrane-anchored isoform, GAD65. J Biol Chem. 1999;274:37200–9. - PubMed
-
- Bonifacio E, Lampasona V, Bernasconi L, Ziegler A-G. Maturation of the humoral autoimmune response to epitopes of GAD in preclinical childhood type 1 diabetes. Diabetes. 2000;49:202–8. - PubMed
-
- Solimena M, Folli F, Denis-Donini S. Autoantibodies to glutamate decarboxylase in a patient with stiff man syndrome, epilepsy and type 1 diabetes mellitus. N Engl J Med. 1988;318:1012–20. - PubMed
-
- Baekkeskov S, Aanstoot HJ, Christgau S, et al. Identification of the 64k autoantigen in insulin-dependent diabetes as the GABA synthesizing enzyme glutamic acid decarboxylase. Nature. 1990;347:151–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

