Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors
- PMID: 12297621
- PMCID: PMC130569
- DOI: 10.1073/pnas.162471999
Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors
Abstract
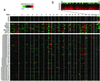
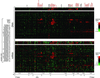
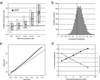
Genomic DNA copy number alterations are key genetic events in the development and progression of human cancers. Here we report a genome-wide microarray comparative genomic hybridization (array CGH) analysis of DNA copy number variation in a series of primary human breast tumors. We have profiled DNA copy number alteration across 6,691 mapped human genes, in 44 predominantly advanced, primary breast tumors and 10 breast cancer cell lines. While the overall patterns of DNA amplification and deletion corroborate previous cytogenetic studies, the high-resolution (gene-by-gene) mapping of amplicon boundaries and the quantitative analysis of amplicon shape provide significant improvement in the localization of candidate oncogenes. Parallel microarray measurements of mRNA levels reveal the remarkable degree to which variation in gene copy number contributes to variation in gene expression in tumor cells. Specifically, we find that 62% of highly amplified genes show moderately or highly elevated expression, that DNA copy number influences gene expression across a wide range of DNA copy number alterations (deletion, low-, mid- and high-level amplification), that on average, a 2-fold change in DNA copy number is associated with a corresponding 1.5-fold change in mRNA levels, and that overall, at least 12% of all the variation in gene expression among the breast tumors is directly attributable to underlying variation in gene copy number. These findings provide evidence that widespread DNA copy number alteration can lead directly to global deregulation of gene expression, which may contribute to the development or progression of cancer.
Figures

References
-
- Kallioniemi A, Kallioniemi O P, Sudar D, Rutovitz D, Gray J W, Waldman F, Pinkel D. Science. 1992;258:818–821. - PubMed
-
- Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi O P. Genes Chromosomes Cancer. 1998;21:177–184. - PubMed
-
- Forozan F, Mahlamaki E H, Monni O, Chen Y, Veldman R, Jiang Y, Gooden G C, Ethier S P, Kallioniemi A, Kallioniemi O P. Cancer Res. 2000;60:4519–4525. - PubMed
-
- Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, Cremer T, Lichter P. Genes Chromosomes Cancer. 1997;20:399–407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

