Rice genome organization: the centromere and genome interactions
- PMID: 12324265
- PMCID: PMC4240384
- DOI: 10.1093/aob/mcf218
Rice genome organization: the centromere and genome interactions
Abstract
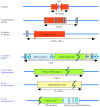
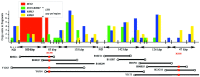
Over the last decade, many varied resources have become available for genome studies in rice. These resources include over 4000 DNA markers, several bacterial artificial chromosome (BAC) libraries, P-1 derived artificial chromosome (PAC) libraries and yeast artificial chromosome (YAC) libraries (genomic DNA clones, filters and end-sequences), retrotransposon tagged lines, and many chemical and irradiated mutant lines. Based on these, high-density genetic maps, cereal comparative maps, YAC and BAC physical maps, and quantitative trait loci (QTL) maps have been constructed, and 93 % of the genome has also been sequenced. These data have revealed key features of the genetic and physical structure of the rice genome and of the evolution of cereal chromosomes. This Botanical Briefing examines aspects of how the rice genome is organized structurally, functionally and evolutionarily. Emphasis is placed on the rice centromere, which is composed of long arrays of centromere-specific repetitive sequences. Differences and similarities amongst various cereal centromeres are detailed. These indicate essential features of centromere function. Another view of various kinds of interactive relationships within and between genomes, which could play crucial roles in genome organization and evolution, is also introduced. Constructed genetic and physical maps indicate duplication of chromosomal segments and spatial association between specific chromosome regions. A genome-wide survey of interactive genetic loci has identified various reproductive barriers that may drive speciation of the rice genome. The significance of these findings in genome organization and evolution is discussed.
Figures
Similar articles
-
Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon.Plant Cell. 2002 Aug;14(8):1691-704. doi: 10.1105/tpc.003079. Plant Cell. 2002. PMID: 12172016 Free PMC article.
-
[Isolation and characterization of the centromeric BAC clones from different genomes in genus Oryza].Yi Chuan. 2007 Jul;29(7):851-8. doi: 10.1360/yc-007-0851. Yi Chuan. 2007. PMID: 17646152 Chinese.
-
Identification and mapping of expressed genes, simple sequence repeats and transposable elements in centromeric regions of rice chromosomes.DNA Res. 2006 Dec 31;13(6):267-74. doi: 10.1093/dnares/dsm001. Epub 2007 Feb 12. DNA Res. 2006. PMID: 17298954
-
From mapping to sequencing, post-sequencing and beyond.Plant Cell Physiol. 2005 Jan;46(1):3-13. doi: 10.1093/pcp/pci503. Epub 2005 Jan 19. Plant Cell Physiol. 2005. PMID: 15659433 Review.
-
Cereal genome analysis using rice as a model.Curr Opin Genet Dev. 1996 Dec;6(6):711-4. doi: 10.1016/s0959-437x(96)80025-6. Curr Opin Genet Dev. 1996. PMID: 8994841 Review.
Cited by
-
New DArT markers for oat provide enhanced map coverage and global germplasm characterization.BMC Genomics. 2009 Jan 21;10:39. doi: 10.1186/1471-2164-10-39. BMC Genomics. 2009. PMID: 19159465 Free PMC article.
-
Genetic diversity assessment and genome-wide association study reveal candidate genes associated with component traits in sweet potato (Ipomoea batatas (L.) Lam).Mol Genet Genomics. 2023 May;298(3):653-667. doi: 10.1007/s00438-023-02007-3. Epub 2023 Mar 21. Mol Genet Genomics. 2023. PMID: 36943475
-
Wheat in vivo RNA structure landscape reveals a prevalent role of RNA structure in modulating translational subgenome expression asymmetry.Genome Biol. 2021 Nov 30;22(1):326. doi: 10.1186/s13059-021-02549-y. Genome Biol. 2021. PMID: 34847934 Free PMC article.
-
Validation of rice genome sequence by optical mapping.BMC Genomics. 2007 Aug 15;8:278. doi: 10.1186/1471-2164-8-278. BMC Genomics. 2007. PMID: 17697381 Free PMC article.
-
Mapping of QTLs associated with yield and related traits under reproductive stage drought stress in rice using SNP linkage map.Mol Biol Rep. 2023 Aug;50(8):6349-6359. doi: 10.1007/s11033-023-08550-x. Epub 2023 Jun 14. Mol Biol Rep. 2023. PMID: 37314604
References
-
- AdamG, Muller JA, Kindle KL.1997. Retrofitting YACs for direct DNA transfer into plant cells. Plant Journal 11: 1349–1358. - PubMed
-
- AntonioBA, Inoue T, Kajiya H, Nagamura Y, Kurata N, Minobe Y, Yano M, Nakagahra M, Sasaki T.1996. Comparison of genetic distance and order of DNA markers in five populations of rice. Genome 39: 946–956. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

