Sequencing and transcriptional analysis of the chlorite dismutase gene of Dechloromonas agitata and its use as a metabolic probe
- PMID: 12324326
- PMCID: PMC126438
- DOI: 10.1128/AEM.68.10.4820-4826.2002
Sequencing and transcriptional analysis of the chlorite dismutase gene of Dechloromonas agitata and its use as a metabolic probe
Abstract
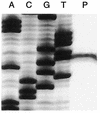
The dismutation of chlorite into chloride and O(2) represents a central step in the reductive pathway of perchlorate that is common to all dissimilatory perchlorate-reducing bacteria and is mediated by a single enzyme, chlorite dismutase. The chlorite dismutase gene cld was isolated and sequenced from the perchlorate-reducing bacterium Dechloromonas agitata strain CKB. Sequence analysis identified an open reading frame of 834 bp that would encode a mature protein with an N-terminal sequence identical to that of the previously purified D. agitata chlorite dismutase enzyme. The predicted translation product of the D. agitata cld gene is a protein of 277 amino acids (aa), including a leader peptide of 26 aa. Primer extension analysis identified a single transcription start site directly downstream of an AT-rich region that could represent the -10 promoter region of the D. agitata cld gene. Northern blot analysis indicated that the cld gene was transcriptionally up-regulated when D. agitata cells were grown in perchlorate-reducing versus aerobic conditions. Slot blot hybridizations with a D. agitata cld probe demonstrated the conservation of the cld gene among perchlorate-reducing bacteria. This study represents the first description of a functional gene associated with microbial perchlorate reduction.
Figures




References
-
- Achenbach, L. A., U. Michaelidou, R. A. Bruce, J. Fryman, and J. D. Coates. 2001. Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int. J. Syst. Evol. Microbiol. 51:527-533. - PubMed
-
- Achenbach, L. A., and J. D. Coates. 2000. Disparity between bacterial phylogeny and physiology. ASM News 66:714-716.
-
- Arkowitz, R., and M. Bassilana. 1994. Protein translocation in Escherichia coli. Biochim. Biophys. Acta 1197:311-343. - PubMed
-
- Aslander, A. 1928. Experiments on the eradication of Canada thistle Cirsium arvense with chlorates and other herbicides. J. Agric. Res. 36:915-935.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

