Diversity and evolution of hydrogenase systems in rhizobia
- PMID: 12324339
- PMCID: PMC126442
- DOI: 10.1128/AEM.68.10.4915-4924.2002
Diversity and evolution of hydrogenase systems in rhizobia
Abstract
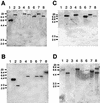
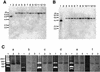
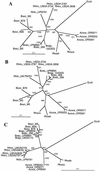
Uptake hydrogenases allow rhizobia to recycle the hydrogen generated in the nitrogen fixation process within the legume nodule. Hydrogenase (hup) systems in Bradyrhizobium japonicum and Rhizobium leguminosarum bv. viciae show highly conserved sequence and gene organization, but important differences exist in regulation and in the presence of specific genes. We have undertaken the characterization of hup gene clusters from Bradyrhizobium sp. (Lupinus), Bradyrhizobium sp. (Vigna), and Rhizobium tropici and Azorhizobium caulinodans strains with the aim of defining the extent of diversity in hup gene composition and regulation in endosymbiotic bacteria. Genomic DNA hybridizations using hupS, hupE, hupUV, hypB, and hoxA probes showed a diversity of intraspecific hup profiles within Bradyrhizobium sp. (Lupinus) and Bradyrhizobium sp. (Vigna) strains and homogeneous intraspecific patterns within R. tropici and A. caulinodans strains. The analysis also revealed differences regarding the possession of hydrogenase regulatory genes. Phylogenetic analyses using partial sequences of hupS and hupL clustered R. leguminosarum and R. tropici hup sequences together with those from B. japonicum and Bradyrhizobium sp. (Lupinus) strains, suggesting a common origin. In contrast, Bradyrhizobium sp. (Vigna) hup sequences diverged from the rest of rhizobial sequences, which might indicate that those organisms have evolved independently and possibly have acquired the sequences by horizontal transfer from an unidentified source.
Figures



References
-
- Albrecht, S. L., R. J. Maier, F. J. Hanus, S. A. Russell, D. W. Emerich, and H. J. Evans. 1979. Hydrogenase in Rhizobium japonicum increases nitrogen fixation by nodulated soybeans. Science 203:1255-1257. - PubMed
-
- Arp, D. J. 1992. Hydrogen recycling in symbiotic bacteria, p. 432-460. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman and Hall, New York, N.Y.
-
- Beringer, J. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

