Sequence-dependent denaturation energetics: A major determinant in amyloid disease diversity
- PMID: 12351683
- PMCID: PMC139904
- DOI: 10.1073/pnas.202495199
Sequence-dependent denaturation energetics: A major determinant in amyloid disease diversity
Abstract
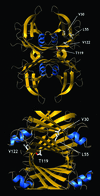
Several misfolding diseases commence when a secreted folded protein encounters a partially denaturing microenvironment, enabling its self assembly into amyloid. Although amyloidosis is modulated by numerous environmental and genetic factors, single point mutations within the amyloidogenic protein can dramatically influence disease phenotype. Mutations that destabilize the native state predispose an individual to disease; however, thermodynamic stability alone does not reliably predict disease severity. Here we show that the rate of transthyretin (TTR) tetramer dissociation required for amyloid formation is strongly influenced by mutation (V30M, L55P, T119M, V122I), with rapid rates exacerbating and slow rates reducing amyloidogenicity. Although these rates are difficult to predict a priori, they notably influence disease penetrance and age of onset. L55P TTR exhibits severe pathology because the tetramer both dissociates quickly and is highly destabilized. Even though V30M and L55P TTR are similarly destabilized, the V30M disease phenotype is milder because V30M dissociates more slowly, even slower than wild type (WT). Although WT and V122I TTR have nearly equivalent tetramer stabilities, V122I cardiomyopathy, unlike WT cardiomyopathy, has nearly complete penetrance-presumably because of its 2-fold increase in dissociation rate. We show that the T119M homotetramer exhibits kinetic stabilization and therefore dissociates exceedingly slowly, likely explaining how it functions to protect V30MT119M compound heterozygotes from disease. An understanding of how mutations influence both the kinetics and thermodynamics of misfolding allows us to rationalize the phenotypic diversity of amyloid diseases, especially when considered in concert with other genetic and environmental data.
Figures
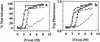
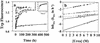


References
-
- Kelly J W. Curr Opin Struct Biol. 1996;6:11–17. - PubMed
-
- Dobson, C. M. (1999) Trends Biochem. Sci. 329–332. - PubMed
-
- Goldberg M S, Lansbury P T., Jr Nat Cell Biol. 2000;2:E115–E119. - PubMed
-
- Fink A L. Folding Des. 1998;3:R9–R23. - PubMed
-
- Uemichi T. Rinsho Kagaku (Nippon Rinsho Kagakkai) 1997;26:74–87.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

