Characterization of multidrug-resistant Escherichia coli isolates associated with nosocomial infections in dogs
- PMID: 12354850
- PMCID: PMC130861
- DOI: 10.1128/JCM.40.10.3586-3595.2002
Characterization of multidrug-resistant Escherichia coli isolates associated with nosocomial infections in dogs
Erratum in
- J Clin Microbiol 2002 Dec;40(12):4806
Abstract
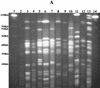
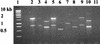
Multidrug-resistant opportunistic pathogens have become endemic to the veterinary hospital environment. Escherichia coli isolates resistant to 12 antibiotics were isolated from two dogs that were housed in the intensive care unit at The University of Georgia Veterinary Teaching Hospital within 48 h of each other. Review of 21 retrospective and prospective hospital-acquired E. coli infections revealed that the isolates had similar antibiotic resistance profiles, characterized by resistance to most cephalosporins, beta-lactams, and the beta-lactamase inhibitor clavulanic acid as well as resistance to tetracycline, spectinomycin, sulfonamides, chloramphenicol, and gentamicin. E. coli isolates with similar resistance profiles were also isolated from the environment in the intensive care unit and surgery wards. Multiple E. coli genetic types were endemic to the hospital environment, with the pulsed-field gel electrophoresis fingerprint identified among E. coli isolates from diseased animals and the hospital environment matching. The extended-spectrum cephalosporin resistance in these nosocomial E. coli isolates was attributed to the cephamycinase-encoding gene, bla(CMY2). Chloramphenicol resistance was due in part to the dissemination of the florfenicol resistance gene, flo, among these isolates. Resistance encoded by both genes was self-transmissible. Although bla(CMY2) and flo were common to the polyclonal, nosocomial E. coli isolates, there was considerable diversity in the genetic compositions of class 1 integrons, especially among isolates belonging to the same genetic type. Two or more integrons were generally present in these isolates. The gene cassettes present within each integron ranged in size from 0.6 to 2.4 kb, although a 1.7-kb gene cassette was the most prevalent. The 1.7-kb gene cassette contained spectinomycin resistance gene aadA5 and trimethoprim resistance gene dfrA17.
Figures




References
-
- Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, J. G. Wells, B. P. Bell, K. D. Greene, J. Lewis, and P. M. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. - PMC - PubMed
-
- Beutin, L. 1999. Escherichia coli as a pathogen in dogs and cats. Vet. Res. 30:285-298. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

