Expression profiling of human renal carcinomas with functional taxonomic analysis
- PMID: 12356337
- PMCID: PMC130042
- DOI: 10.1186/1471-2105-3-26
Expression profiling of human renal carcinomas with functional taxonomic analysis
Abstract
Background: Molecular characterization has contributed to the understanding of the inception, progression, treatment and prognosis of cancer. Nucleic acid array-based technologies extend molecular characterization of tumors to thousands of gene products. To effectively discriminate between tumor sub-types, reliable laboratory techniques and analytic methods are required.
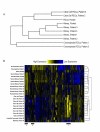
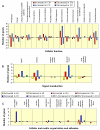
Results: We derived mRNA expression profiles from 21 human tissue samples (eight normal kidneys and 13 kidney tumors) and two pooled samples using the Affymetrix GeneChip platform. A panel of ten clustering algorithms combined with four data pre-processing methods identified a consensus cluster dendrogram in 18 of 40 analyses and of these 16 used a logarithmic transformation. Within the consensus dendrogram the expression profiles of the samples grouped according to tissue type; clear cell and chromophobe carcinomas displayed distinctly different gene expression patterns. By using a rigorous statistical selection based method we identified 355 genes that showed significant (p < 0.001) gene expression changes in clear cell renal carcinomas compared to normal kidney. These genes were classified with a tool to conceptualize expression patterns called "Functional Taxonomy". Each tumor type had a distinct "signature," with a high number of genes in the categories of Metabolism, Signal Transduction, and Cellular and Matrix Organization and Adhesion.
Conclusions: Affymetrix GeneChip profiling differentiated clear cell and chromophobe carcinomas from one another and from normal kidney cortex. Clustering methods that used logarithmic transformation of data sets produced dendrograms consistent with the sample biology. Functional taxonomy provided a practical approach to the interpretation of gene expression data.
Figures


References
-
- Fitzgibbons PL, Page DL, Weaver D, Thor AD, Allred DC, Clark GM, Ruby SG, O'Malley F, Simpson JF, Connolly JL. Prognostic factors in breast cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124:966–978. - PubMed
-
- Kovacs G. Molecular genetics of human renal cell tumors. Nephrol Dial Transplant. 1996;11(Suppl 6):62–65. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

