The cross-reactive calcium-binding pollen allergen, Phl p 7, reveals a novel dimer assembly
- PMID: 12356717
- PMCID: PMC129048
- DOI: 10.1093/emboj/cdf526
The cross-reactive calcium-binding pollen allergen, Phl p 7, reveals a novel dimer assembly
Abstract
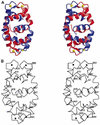
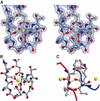
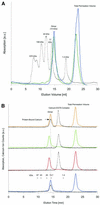
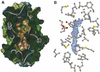
The timothy grass pollen allergen Phl p 7 assembles most of the IgE epitopes of a novel family of 2 EF-hand calcium-binding proteins and therefore represents a diagnostic marker allergen and vaccine candidate for immunotherapy. Here we report the first three-dimensional structure of a representative of the 2 EF-hand allergen family, Phl p 7, in the calcium-bound form. The protein occurs as a novel dimer assembly with unique features: in contrast to well known EF-hand proteins such as calmodulin, parvalbumin or the S100 proteins, Phl p 7 adopts an extended conformation. Two protein monomers assemble in a head-to-tail arrangement with domain-swapped EF-hand pairing. The intertwined dimer adopts a barrel-like structure with an extended hydrophobic cavity providing a ligand-binding site. Calcium binding acts as a conformational switch between an open and a closed dimeric form of Phl p 7. These findings are interesting in the context of lipid- and calcium-dependent pollen tube growth. Furthermore, the structure of Phl p 7 allows for the rational development of vaccine strategies for treatment of sensitized allergic patients.
Figures

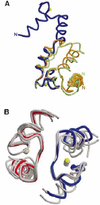

Similar articles
-
Solution structure, dynamics, and hydrodynamics of the calcium-bound cross-reactive birch pollen allergen Bet v 4 reveal a canonical monomeric two EF-hand assembly with a regulatory function.J Mol Biol. 2004 Mar 5;336(5):1141-57. doi: 10.1016/j.jmb.2003.12.070. J Mol Biol. 2004. PMID: 15037075
-
Calcium-dependent immunoglobulin E recognition of the apo- and calcium-bound form of a cross-reactive two EF-hand timothy grass pollen allergen, Phl p 7.FASEB J. 1999 May;13(8):843-56. doi: 10.1096/fasebj.13.8.843. FASEB J. 1999. PMID: 10224228
-
Generation of an allergy vaccine by disruption of the three-dimensional structure of the cross-reactive calcium-binding allergen, Phl p 7.J Immunol. 2004 May 1;172(9):5684-92. doi: 10.4049/jimmunol.172.9.5684. J Immunol. 2004. PMID: 15100313
-
Molecular characterization of timothy grass pollen group V allergens.Int Arch Allergy Immunol. 1995 May-Jun;107(1-3):242-4. doi: 10.1159/000236991. Int Arch Allergy Immunol. 1995. PMID: 7542075 Review.
-
Calcium-binding proteins and their role in allergic diseases.Immunol Allergy Clin North Am. 2007 Feb;27(1):29-44. doi: 10.1016/j.iac.2006.10.003. Immunol Allergy Clin North Am. 2007. PMID: 17276877 Review.
Cited by
-
Structure of a patient-derived antibody in complex with allergen reveals simultaneous conventional and superantigen-like recognition.Proc Natl Acad Sci U S A. 2018 Sep 11;115(37):E8707-E8716. doi: 10.1073/pnas.1806840115. Epub 2018 Aug 27. Proc Natl Acad Sci U S A. 2018. PMID: 30150373 Free PMC article.
-
Structure of allergens and structure based epitope predictions.Methods. 2014 Mar 1;66(1):3-21. doi: 10.1016/j.ymeth.2013.07.024. Epub 2013 Jul 23. Methods. 2014. PMID: 23891546 Free PMC article. Review.
-
Panallergens and their impact on the allergic patient.Allergy Asthma Clin Immunol. 2010 Jan 18;6(1):1. doi: 10.1186/1710-1492-6-1. Allergy Asthma Clin Immunol. 2010. PMID: 20298513 Free PMC article.
-
Analysis of Pollen Allergens in Lily by Transcriptome and Proteome Data.Int J Mol Sci. 2019 Nov 24;20(23):5892. doi: 10.3390/ijms20235892. Int J Mol Sci. 2019. PMID: 31771269 Free PMC article.
-
Effect of tomato variety, cultivation, climate and processing on Sola l 4, an allergen from Solanum lycopersicum.PLoS One. 2018 Jun 14;13(6):e0197971. doi: 10.1371/journal.pone.0197971. eCollection 2018. PLoS One. 2018. PMID: 29902173 Free PMC article.
References
-
- Babu Y.S., Bugg,C.E. and Cook,W.J. (1988) Structure of calmodulin refined at 2.2 Å resolution. J. Mol. Biol., 204, 191–204. - PubMed
-
- Batanero E., Villalba,M., Ledesma,A., Puente,X.S. and Rodriguez,R. (1996) Ole e 3, an olive-tree allergen, belongs to a widespread family of pollen proteins. Eur. J. Biochem., 241, 772–778. - PubMed
-
- Brewbaker J.L. and Kwack,B.H. (1963) The essential role of calcium ion in pollen germination and pollen tube growth. Am. J. Bot., 50, 859–865.
-
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

