The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus
- PMID: 12356718
- PMCID: PMC129051
- DOI: 10.1093/emboj/cdf529
The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus
Abstract
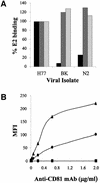
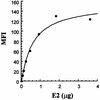
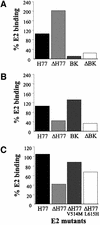
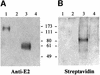
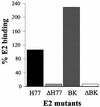
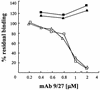
We discovered that the hepatitis C virus (HCV) envelope glycoprotein E2 binds to human hepatoma cell lines independently of the previously proposed HCV receptor CD81. Comparative binding studies using recombinant E2 from the most prevalent 1a and 1b genotypes revealed that E2 recognition by hepatoma cells is independent from the viral isolate, while E2-CD81 interaction is isolate specific. Binding of soluble E2 to human hepatoma cells was impaired by deletion of the hypervariable region 1 (HVR1), but the wild-type phenotype was recovered by introducing a compensatory mutation reported previously to rescue infectivity of an HVR1-deleted HCV infectious clone. We have identified the receptor responsible for E2 binding to human hepatic cells as the human scavenger receptor class B type I (SR-BI). E2-SR-BI interaction is very selective since neither mouse SR-BI nor the closely related human scavenger receptor CD36, were able to bind E2. Finally, E2 recognition by SR-BI was competed out in an isolate-specific manner both on the hepatoma cell line and on the human SR-BI-transfected cell line by an anti-HVR1 monoclonal antibody.
Figures



References
-
- Acton S.L., Scherer,P.E., Lodish,H.F. and Krieger,M. (1994) Expression cloning of SR-BI, a CD36-related class B scavenger receptor. J. Biol. Chem., 269, 21003–21009. - PubMed
-
- Acton S., Rigotti,A., Landschulz,K.T., Xu,S., Hobbs,H.H. and Krieger,M. (1996) Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science, 271, 518–520. - PubMed
-
- Allander T., Forns,X., Emerson,S.U., Purcell,R.H. and Bukh,J. (2000) Hepatitis C virus envelope protein E2 binds to CD81 of tamarins. Virology, 277, 358–367. - PubMed
-
- Babitt J., Trigatti,B., Rigotti,A., Smart,E.J., Anderson,R.G., Xu,S. and Krieger,M. (1997) Murine SR-BI, a high density lipoprotein receptor that mediates selective lipid uptake, is N-glycosylated and fatty acylated and colocalizes with plasma membrane caveolae. J. Biol. Chem., 272, 13242–13249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

