The Drosophila BRM complex facilitates global transcription by RNA polymerase II
- PMID: 12356740
- PMCID: PMC129039
- DOI: 10.1093/emboj/cdf517
The Drosophila BRM complex facilitates global transcription by RNA polymerase II
Abstract
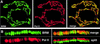
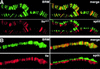
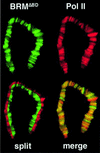
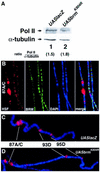
Drosophila brahma (brm) encodes the ATPase subunit of a 2 MDa complex that is related to yeast SWI/SNF and other chromatin-remodeling complexes. BRM was identified as a transcriptional activator of Hox genes required for the specification of body segment identities. To clarify the role of the BRM complex in the transcription of other genes, we examined its distribution on larval salivary gland polytene chromosomes. The BRM complex is associated with nearly all transcriptionally active chromatin in a pattern that is generally non-overlapping with that of Polycomb, a repressor of Hox gene transcription. Reduction of BRM function dramatically reduces the association of RNA polymerase II with salivary gland chromosomes. A few genes, such as induced heat shock loci, are not associated with the BRM complex; transcription of these genes is not compromised by loss of BRM function. The distribution of the BRM complex thus correlates with a dependence on BRM for gene activity. These data suggest that the chromatin remodeling activity of the BRM complex plays a general role in facilitating transcription by RNA polymerase II.
Figures





References
-
- Armstrong J.A., Bieker,J.J. and Emerson,B.M. (1998) A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell, 95, 93–104. - PubMed
-
- Boyer L.A. and Peterson,C.L. (2000) Actin-related proteins (Arps): conformational switches for chromatin-remodeling machines? BioEssays, 22, 666–672. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

