Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array
- PMID: 12356743
- PMCID: PMC129033
- DOI: 10.1093/emboj/cdf511
Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array
Abstract
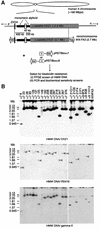
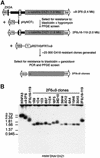
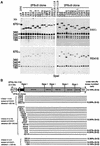
Dissection of human centromeres is difficult because of the lack of landmarks within highly repeated DNA. We have systematically manipulated a single human X centromere generating a large series of deletion derivatives, which have been examined at four levels: linear DNA structure; the distribution of constitutive centromere proteins; topoisomerase IIalpha cleavage activity; and mitotic stability. We have determined that the human X major alpha-satellite locus, DXZ1, is asymmetrically organized with an active subdomain anchored approximately 150 kb in from the Xp-edge. We demonstrate a major site of topoisomerase II cleavage within this domain that can shift if juxtaposed with a telomere, suggesting that this enzyme recognizes an epigenetic determinant within the DXZ1 chromatin. The observation that the only part of the DXZ1 locus shared by all deletion derivatives is a highly restricted region of <50 kb, which coincides with the topo isomerase II cleavage site, together with the high levels of cleavage detected, identify topoisomerase II as a major player in centromere biology.
Figures


References
-
- Bayne R.A.L., Broccoli,D., Taggart,M.H., Thomson,E.J., Farr,C.J. and Cooke,H.J. (1994) Sandwiching of a gene within 12 kb of a functional telomere and α-satellite does not result in silencing. Hum. Mol. Genet., 3, 539–546. - PubMed
-
- Bernard P., Maure,J.F., Partridge,J.F., Genier,S., Javerzat,J.P. and Allshire,R.C. (2001) Requirement of heterochromatin for cohesion at centromeres. Science, 294, 2539–2542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

