An extracellular site on tetraspanin CD151 determines alpha 3 and alpha 6 integrin-dependent cellular morphology
- PMID: 12356873
- PMCID: PMC2173251
- DOI: 10.1083/jcb.200204056
An extracellular site on tetraspanin CD151 determines alpha 3 and alpha 6 integrin-dependent cellular morphology
Abstract
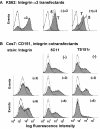
The alpha 3 beta 1 integrin shows strong, stoichiometric, direct lateral association with the tetraspanin CD151. As shown here, an extracellular CD151 site (QRD(194-196)) is required for strong (i.e., Triton X-100-resistant) alpha 3 beta 1 association and for maintenance of a key CD151 epitope (defined by monoclonal antibody TS151r) that is blocked upon alpha 3 integrin association. Strong CD151 association with integrin alpha 6 beta 1 also required the QRD(194-196) site and masked the TS151r epitope. For both alpha 3 and alpha 6 integrins, strong QRD/TS151r-dependent CD151 association occurred early in biosynthesis and involved alpha subunit precursor forms. In contrast, weaker associations of CD151 with itself, integrins, or other tetraspanins (Triton X-100-sensitive but Brij 96-resistant) were independent of the QRD/TS151r site, occurred late in biosynthesis, and involved mature integrin subunits. Presence of the CD151-QRD(194-196)-->INF mutant disrupted alpha 3 and alpha 6 integrin-dependent formation of a network of cellular cables by Cos7 or NIH3T3 cells on basement membrane Matrigel and markedly altered cell spreading. These results provide definitive evidence that strong lateral CD151-integrin association is functionally important, identify CD151 as a key player during alpha 3 and alpha 6 integrin-dependent matrix remodeling and cell spreading, and support a model of CD151 as a transmembrane linker between extracellular integrin domains and intracellular cytoskeleton/signaling molecules.
Figures










References
-
- Anton, E.S., J.A. Kreidberg, and P. Rakic. 1999. Distinct functions of alpha3 and alpha(v) integrin receptors in neuronal migration and laminar organization of the cerebral cortex. Neuron. 22:277–289. - PubMed
-
- Belkin, A.M., and M.A. Stepp. 2000. Integrins as receptors for laminins. Microsc. Res. Tech. 51:280–301. - PubMed
-
- Berditchevski, F. 2001. Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 114:4143–4151. - PubMed
-
- Berditchevski, F., G. Bazzoni, and M.E. Hemler. 1995. Specific association of CD63 with the VLA-3 and VLA-6 integrins. J. Biol. Chem. 270:17784–17790. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

