Selenomethionine regulation of p53 by a ref1-dependent redox mechanism
- PMID: 12357032
- PMCID: PMC137920
- DOI: 10.1073/pnas.212319799
Selenomethionine regulation of p53 by a ref1-dependent redox mechanism
Abstract
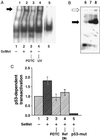
The cancer chemopreventive properties of selenium compounds are well documented, yet little is known of the mechanism(s) by which these agents inhibit carcinogenesis. We show that selenium in the form of selenomethionine (SeMet) can activate the p53 tumor suppressor protein by a redox mechanism that requires the redox factor Ref1. Assays to measure direct reduction/oxidation of p53 showed a SeMet-dependent response that was blocked by a dominant-negative Ref1. By using a peptide containing only p53 cysteine residues 275 and 277, we demonstrate the importance of these residues in the SeMet-induced response. SeMet induced sequence-specific DNA binding and transactivation by p53. Finally, cellular responses to SeMet were determined in mouse embryo fibroblasts wild-type or null for p53 genes. The evidence suggests that the DNA repair branch of the p53 pathway was activated. The central relevance of DNA repair to cancer prevention is discussed.
Figures
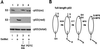
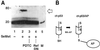


Comment in
-
New careers for antioxidants.Proc Natl Acad Sci U S A. 2002 Oct 29;99(22):13969-71. doi: 10.1073/pnas.232574399. Epub 2002 Oct 21. Proc Natl Acad Sci U S A. 2002. PMID: 12391310 Free PMC article. Review. No abstract available.
References
-
- Ip C. (1981) Cancer Res. 41, 4386-4390. - PubMed
-
- Reddy B. S., Sugie, S., Maryyama, H., el-Bayoumy, K. & Marra, P. (1987) Cancer Res. 47, 5901-5904. - PubMed
-
- Nelson M. A., Porterfield, B. W., Jacobs, E. T. & Clark, L. C. (1999) Semin. Urol. Oncol. 17, 91-96. - PubMed
-
- Lu J., Jiang, C., Kaeck, M., Ganther, H., Vadhanavikit, S., Ip, C. & Thompson, H. J. (1995) Biochem. Pharmacol. 50, 213-219. - PubMed
-
- Sinha R., Said, T. K. & Medina, D. (1996) Cancer Lett. (Shannon, Irel.) 107, 277-284. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

