Solution structure and dynamics of the outer membrane enzyme PagP by NMR
- PMID: 12357033
- PMCID: PMC129713
- DOI: 10.1073/pnas.212344499
Solution structure and dynamics of the outer membrane enzyme PagP by NMR
Abstract
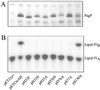
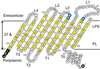
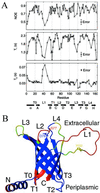
The bacterial outer membrane enzyme PagP transfers a palmitate chain from a phospholipid to lipid A. In a number of pathogenic Gram-negative bacteria, PagP confers resistance to certain cationic antimicrobial peptides produced during the host innate immune response. The global fold of Escherichia coli PagP was determined in both dodecylphosphocholine and n-octyl-beta-d-glucoside detergent micelles using solution NMR spectroscopy. PagP consists of an eight-stranded anti-parallel beta-barrel preceded by an amphipathic alpha helix. The beta-barrel is well defined, whereas NMR relaxation measurements reveal considerable mobility in the loops connecting individual beta-strands. Three amino acid residues critical for enzymatic activity localize to extracellular loops near the membrane interface, positioning them optimally to interact with the polar headgroups of lipid A. Hence, the active site of PagP is situated on the outer surface of the outer membrane. Because the phospholipids that donate palmitate in the enzymatic reaction are normally found only in the inner leaflet of the outer membrane, PagP activity may depend on the aberrant migration of phospholipids into the outer leaflet. This finding is consistent with an emerging paradigm for outer membrane enzymes in providing an adaptive response toward disturbances in the outer membrane.
Figures



Similar articles
-
The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis.Mol Microbiol. 2005 Aug;57(4):900-12. doi: 10.1111/j.1365-2958.2005.04711.x. Mol Microbiol. 2005. PMID: 16091033 Review.
-
The N-terminal helix is a post-assembly clamp in the bacterial outer membrane protein PagP.J Mol Biol. 2007 Oct 26;373(3):529-40. doi: 10.1016/j.jmb.2007.07.072. Epub 2007 Aug 15. J Mol Biol. 2007. PMID: 17868697 Free PMC article.
-
Distinct Structural Elements Govern the Folding, Stability, and Catalysis in the Outer Membrane Enzyme PagP.Biochemistry. 2016 Sep 6;55(35):4960-70. doi: 10.1021/acs.biochem.6b00678. Epub 2016 Aug 24. Biochemistry. 2016. PMID: 27525547
-
Enzymology of lipid A palmitoylation in bacterial outer membranes.J Endotoxin Res. 2004;10(2):107-12. doi: 10.1179/096805104225004004. J Endotoxin Res. 2004. PMID: 15120001 Review.
-
One membrane protein, two structures and six environments: a comparative molecular dynamics simulation study of the bacterial outer membrane protein PagP.Mol Membr Biol. 2009 May;26(4):205-14. doi: 10.1080/09687680902788967. Epub 2009 Mar 10. Mol Membr Biol. 2009. PMID: 19280380
Cited by
-
The Raetz pathway for lipid A biosynthesis: Christian Rudolf Hubert Raetz, MD PhD, 1946–2011.J Lipid Res. 2011 Nov;52(11):1857-1860. doi: 10.1194/jlr.e020701. J Lipid Res. 2011. PMID: 22106472 Free PMC article. No abstract available.
-
Structure-Function Characterization of the Conserved Regulatory Mechanism of the Escherichia coli M48 Metalloprotease BepA.J Bacteriol. 2020 Dec 18;203(2):e00434-20. doi: 10.1128/JB.00434-20. Print 2020 Dec 18. J Bacteriol. 2020. PMID: 33106348 Free PMC article.
-
The bacterial cell envelope.Cold Spring Harb Perspect Biol. 2010 May;2(5):a000414. doi: 10.1101/cshperspect.a000414. Epub 2010 Apr 14. Cold Spring Harb Perspect Biol. 2010. PMID: 20452953 Free PMC article. Review.
-
Mapping membrane protein backbone dynamics: a comparison of site-directed spin labeling with NMR 15N-relaxation measurements.Biophys J. 2014 Oct 7;107(7):1697-702. doi: 10.1016/j.bpj.2014.08.018. Biophys J. 2014. PMID: 25296323 Free PMC article.
-
Outer membrane proteins: comparing X-ray and NMR structures by MD simulations in lipid bilayers.Eur Biophys J. 2008 Feb;37(2):131-41. doi: 10.1007/s00249-007-0185-8. Epub 2007 Jun 6. Eur Biophys J. 2008. PMID: 17551722
References
-
- Aderem A, Ulevitch R J. Nature. 2000;406:782–787. - PubMed
-
- Christ W J, Asano O, Robidoux A L, Perez M, Wang Y, Dubuc G R, Gavin W E, Hawkins L D, McGuinness P D, Mullarkey M A, et al. Science. 1995;268:80–83. - PubMed
-
- Guo L, Lim K B, Gunn J S, Bainbridge B, Darveau R P, Hackett M, Miller S L. Science. 1997;276:250–253. - PubMed
-
- Ernst R K, Yi E C, Guo L, Lim K B, Burns J L, Hackett M, Miller S I. Science. 1999;286:1561–1565. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

