Interaction of dihydrofolate reductase with methotrexate: ensemble and single-molecule kinetics
- PMID: 12359872
- PMCID: PMC129699
- DOI: 10.1073/pnas.172501499
Interaction of dihydrofolate reductase with methotrexate: ensemble and single-molecule kinetics
Abstract
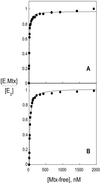
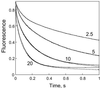
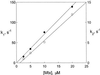
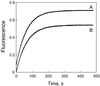
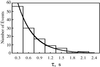
The thermodynamics and kinetics of the interaction of dihydrofolate reductase (DHFR) with methotrexate have been studied by using fluorescence, stopped-flow, and single-molecule methods. DHFR was modified to permit the covalent addition of a fluorescent molecule, Alexa 488, and a biotin at the N terminus of the molecule. The fluorescent molecule was placed on a protein loop that closes over methotrexate when binding occurs, thus causing a quenching of the fluorescence. The biotin was used to attach the enzyme in an active form to a glass surface for single-molecule studies. The equilibrium dissociation constant for the binding of methotrexate to the enzyme is 9.5 nM. The stopped-flow studies revealed that methotrexate binds to two different conformations of the enzyme, and the association and dissociation rate constants were determined. The single-molecule investigation revealed a conformational change in the enzyme-methotrexate complex that was not observed in the stopped-flow studies. The ensemble averaged rate constants for this conformation change in both directions is about 2-4 s(-1) and is attributed to the opening and closing of the enzyme loop over the bound methotrexate. Thus the mechanism of methotrexate binding to DHFR involves multiple steps and protein conformational changes.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

