The human L-threonine 3-dehydrogenase gene is an expressed pseudogene
- PMID: 12361482
- PMCID: PMC131051
- DOI: 10.1186/1471-2156-3-18
The human L-threonine 3-dehydrogenase gene is an expressed pseudogene
Abstract
Background: L-threonine is an indispensable amino acid. One of the major L-threonine degradation pathways is the conversion of L-threonine via 2-amino-3-ketobutyrate to glycine. L-threonine dehydrogenase (EC 1.1.1.103) is the first enzyme in the pathway and catalyses the reaction: L-threonine + NAD+ = 2-amino-3-ketobutyrate + NADH. The murine and porcine L-threonine dehydrogenase genes (TDH) have been identified previously, but the human gene has not been identified.
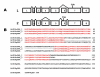
Results: The human TDH gene is located at 8p23-22 and has 8 exons spanning 10 kb that would have been expected to encode a 369 residue ORF. However, 2 cDNA TDH transcripts encode truncated proteins of 157 and 230 residues. These truncated proteins are the result of 3 mutations within the gene. There is a SNP, A to G, present in the genomic DNA sequence of some individuals which results in the loss of the acceptor splice site preceding exon 4. The acceptor splice site preceding exon 6 was lost in all 23 individuals genotyped and there is an in-frame stop codon in exon 6 (CGA to TGA) resulting in arginine-214 being replaced by a stop codon. These truncated proteins would be non-functional since they have lost part of the NAD+ binding motif and the COOH terminal domain that is thought to be involved in binding L-threonine. TDH mRNA was present in all tissues examined.
Conclusions: The human L-threonine 3-dehydrogenase gene is an expressed pseudogene having lost the splice acceptor site preceding exon 6 and codon arginine-214 (CGA) is mutated to a stop codon (TGA).
Figures








References
-
- Dabos KJ, Nelson LJ, Bradnock TJ, Parkinson JA, Sadler IH, Hayes PC, Plevris JN. The simulated microgravity environment maintains key metabolic functions and promotes aggregation of primary porcine hepatocytes. Biochim Biophys Acta. 2001;1526:119–130. doi: 10.1016/S0304-4165(01)00097-6. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

