Combinatorial mutagenesis to restrict amino acid usage in an enzyme to a reduced set
- PMID: 12361984
- PMCID: PMC129711
- DOI: 10.1073/pnas.222243999
Combinatorial mutagenesis to restrict amino acid usage in an enzyme to a reduced set
Abstract
We developed an effective strategy to restrict the amino acid usage in a relatively large protein to a reduced set with conservation of its in vivo function. The 213-residue Escherichia coli orotate phosphoribosyltransferase was subjected to 22 cycles of segment-wise combinatorial mutagenesis followed by 6 cycles of site-directed random mutagenesis, both coupled with a growth-related phenotype selection. The enzyme eventually tolerated 73 amino acid substitutions: In the final variant, 9 amino acid types (A, D, G, L, P, R, T, V, and Y) occupied 188 positions (88%), and none of 7 amino acid types (C, H, I, M, N, Q, and W) appeared. Therefore, the catalytic function associated with a relatively large protein may be achieved with a subset of the 20 amino acid. The converged sequence also implies simpler constituents for proteins in the early stage of evolution.
Figures
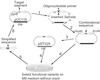


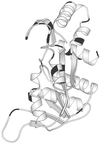
References
-
- Regan L, DeGrado W F. Science. 1988;241:976–978. - PubMed
-
- Kamtekar S, Schiffer J M, Xiong H, Babik J M, Hecht M H. Science. 1993;262:1680–1685. - PubMed
-
- Schafmeister C E, LaPorte S L, Miercke L J, Stroud R M. Nat Struct Biol. 1997;4:1039–1046. - PubMed
-
- Riddle D S, Santiago J V, Bray-Hall S T, Doshi N, Grantcharova V P, Yi Q, Baker D. Nat Struct Biol. 1997;4:805–809. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

