Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance
- PMID: 12364374
- PMCID: PMC126860
- DOI: 10.1128/CMR.15.4.647-679.2002
Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance
Abstract
Infections have been the major cause of disease throughout the history of human populations. With the introduction of antibiotics, it was thought that this problem should disappear. However, bacteria have been able to evolve to become antibiotic resistant. Nowadays, a proficient pathogen must be virulent, epidemic, and resistant to antibiotics. Analysis of the interplay among these features of bacterial populations is needed to predict the future of infectious diseases. In this regard, we have reviewed the genetic linkage of antibiotic resistance and bacterial virulence in the same genetic determinants as well as the cross talk between antibiotic resistance and virulence regulatory circuits with the aim of understanding the effect of acquisition of resistance on bacterial virulence. We also discuss the possibility that antibiotic resistance and bacterial virulence might prevail as linked phenotypes in the future. The novel situation brought about by the worldwide use of antibiotics is undoubtedly changing bacterial populations. These changes might alter the properties of not only bacterial pathogens, but also the normal host microbiota. The evolutionary consequences of the release of antibiotics into the environment are largely unknown, but most probably restoration of the microbiota from the preantibiotic era is beyond our current abilities.
Figures
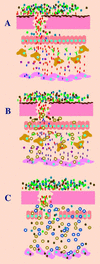


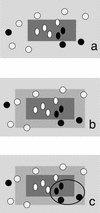

References
-
- Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. - PubMed
-
- Ainsa, J. A., E. Perez, V. Pelicic, F. X. Berthet, B. Gicquel, and C. Martin. 1997. Aminoglycoside 2′-N-acetyltransferase genes are universally present in mycobacteria: characterization of the aac(2′)-Ic gene from Mycobacterium tuberculosis and the aac(2′)-Id gene from Mycobacterium smegmatis. Mol. Microbiol. 24:431-441. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

