RNA canonical and non-canonical base pairing types: a recognition method and complete repertoire
- PMID: 12364604
- PMCID: PMC140540
- DOI: 10.1093/nar/gkf540
RNA canonical and non-canonical base pairing types: a recognition method and complete repertoire
Abstract
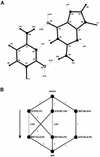
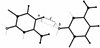
The problem of systematic and objective identification of canonical and non-canonical base pairs in RNA three-dimensional (3D) structures was studied. A probabilistic approach was applied, and an algorithm and its implementation in a computer program that detects and analyzes all the base pairs contained in RNA 3D structures were developed. The algorithm objectively distinguishes among canonical and non-canonical base pairing types formed by three, two and one hydrogen bonds (H-bonds), as well as those containing bifurcated and C-H.X...H-bonds. The nodes of a bipartite graph are used to encode the donor and acceptor atoms of a 3D structure. The capacities of the edges correspond to probabilities computed from the geometry of the donor and acceptor groups to form H-bonds. The maximum flow from donors to acceptors directly identifies base pairs and their types. A complete repertoire of base pairing types was built from the detected H-bonds of all X-ray crystal structures of a resolution of 3.0 A or better, including the large and small ribosomal subunits. The base pairing types are labeled using an extension of the nomenclature recently introduced by Leontis and Westhof. The probabilistic method was implemented in MC-Annotate, an RNA structure analysis computer program used to determine the base pairing parameters of the 3D modeling system MC-Sym.
Figures







References
-
- Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz.T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. - PubMed
-
- Wimberly B.T., Brodersen,D.E., Clemons,W.M.,Jr, Morgan-Warren,R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. - PubMed
-
- Lemieux S., Oldziej,S. and Major,F. (1998) Nucleic acids: qualitative modeling. In Allinger,N.L., Clark,T., Gasteiger,J., Kollman,P.A., Schaefer,H.F. and Schreiner,P.R. (eds), Encyclopedia of Computational Chemistry. John Wiley & Sons, West Sussex, UK.

