Fragile X-related protein and VIG associate with the RNA interference machinery
- PMID: 12368260
- PMCID: PMC187452
- DOI: 10.1101/gad.1025202
Fragile X-related protein and VIG associate with the RNA interference machinery
Abstract
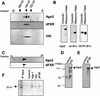
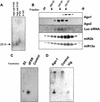
RNA interference (RNAi) is a flexible gene silencing mechanism that responds to double-stranded RNA by suppressing homologous genes. Here, we report the characterization of RNAi effector complexes (RISCs) that contain small interfering RNAs and microRNAs (miRNAs). We identify two putative RNA-binding proteins, the Drosophila homolog of the fragile X mental retardation protein (FMRP), dFXR, and VIG (Vasa intronic gene), through their association with RISC. FMRP, the product of the human fragile X locus, regulates the expression of numerous mRNAs via an unknown mechanism. The possibility that dFXR, and potentially FMRP, use, at least in part, an RNAi-related mechanism for target recognition suggests a potentially important link between RNAi and human disease.
Figures




References
-
- Bardoni B, Mandel JL. Advances in understanding of fragile X pathogenesis and FMRP function, and in identification of X linked mental retardation genes. Curr Opin Genet Dev. 2002;12:284–293. - PubMed
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001a;409:363–366. - PubMed
-
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
