Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM)
- PMID: 12368332
- PMCID: PMC136654
- DOI: 10.1128/jvi.76.21.10894-10904.2002
Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM)
Abstract
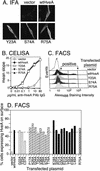
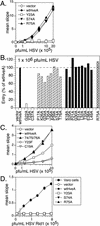
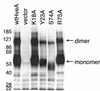
Binding of herpes simplex virus (HSV) envelope glycoprotein D (gD) to a cell surface receptor is an essential step of virus entry. We recently determined the crystal structure of gD bound to one receptor, HveA. HveA is a member of the tumor necrosis factor receptor family and contains four characteristic cysteine-rich domains (CRDs). The first two CRDs of HveA are necessary and sufficient for gD binding. The structure of the gD-HveA complex reveals that 17 amino acids in HveA CRD1 and 4 amino acids in HveA CRD2 directly contact gD. To determine the contribution of these 21 HveA residues to virus entry, we constructed forms of HveA mutated in each of these contact residues. We determined the ability of the mutant proteins to bind gD, facilitate virus entry, and form HveA oligomers. Our results point to a binding hot spot centered around HveA-Y23, a residue that protrudes into a crevice on the surface of gD. Both the hydroxyl group and phenyl group of HveA-Y23 contribute to HSV entry. Our results also suggest that an intermolecular beta-sheet formed between gD and HveA residues 35 to 37 contributes to binding and that a C37-C19 disulfide bond in CRD1 is a critical component of HveA structure necessary for gD binding. The results argue that CRD2 is required for gD binding mainly to provide structural support for a gD binding site in CRD1. Only one mutant, HveA-R75A, exhibited enhanced gD binding. While some mutations influenced complex formation, the majority did not affect HSV entry, suggesting that most contact residues contribute to HveA receptor function collectively rather than individually. This structure-based dissection of the gD-HveA binding site highlights the contribution of key residues within HveA to gD binding and HSV entry and defines a target region for the design of small-molecule inhibitors.
Figures



Similar articles
-
Structure-based mutagenesis of herpes simplex virus glycoprotein D defines three critical regions at the gD-HveA/HVEM binding interface.J Virol. 2003 Jul;77(14):8127-40. doi: 10.1128/jvi.77.14.8127-8140.2003. J Virol. 2003. PMID: 12829851 Free PMC article.
-
Localization of the gD-binding region of the human herpes simplex virus receptor, HveA.J Virol. 2001 Jan;75(1):171-80. doi: 10.1128/JVI.75.1.171-180.2001. J Virol. 2001. PMID: 11119586 Free PMC article.
-
Crystallization and preliminary diffraction studies of the ectodomain of the envelope glycoprotein D from herpes simplex virus 1 alone and in complex with the ectodomain of the human receptor HveA.Acta Crystallogr D Biol Crystallogr. 2002 May;58(Pt 5):836-8. doi: 10.1107/s0907444902001270. Epub 2002 Apr 26. Acta Crystallogr D Biol Crystallogr. 2002. PMID: 11976496
-
The role of herpes simplex virus glycoproteins in the virus replication cycle.Acta Virol. 1998 Apr;42(2):103-18. Acta Virol. 1998. PMID: 9770079 Review.
-
Different receptors binding to distinct interfaces on herpes simplex virus gD can trigger events leading to cell fusion and viral entry.Virology. 2006 Jan 5;344(1):17-24. doi: 10.1016/j.virol.2005.09.016. Virology. 2006. PMID: 16364731 Review.
Cited by
-
Herpes simplex virus glycoprotein B binds to cell surfaces independently of heparan sulfate and blocks virus entry.J Virol. 2005 Sep;79(18):11588-97. doi: 10.1128/JVI.79.18.11588-11597.2005. J Virol. 2005. PMID: 16140736 Free PMC article.
-
NF-kappaB and virus infection: who controls whom.EMBO J. 2003 Jun 2;22(11):2552-60. doi: 10.1093/emboj/cdg267. EMBO J. 2003. PMID: 12773372 Free PMC article. Review.
-
The domains of glycoprotein D required to block apoptosis induced by herpes simplex virus 1 are largely distinct from those involved in cell-cell fusion and binding to nectin1.J Virol. 2003 Mar;77(6):3759-67. doi: 10.1128/jvi.77.6.3759-3767.2003. J Virol. 2003. PMID: 12610150 Free PMC article.
-
Mutations in herpes simplex virus gD protein affect receptor binding by different molecular mechanisms.J Mol Model. 2014 Apr;20(4):2192. doi: 10.1007/s00894-014-2192-x. Epub 2014 Mar 20. J Mol Model. 2014. PMID: 24647818
-
Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops.J Virol. 2009 Jul;83(13):6825-36. doi: 10.1128/JVI.00301-09. Epub 2009 Apr 15. J Virol. 2009. PMID: 19369321 Free PMC article.
References
-
- Banner, D. W., A. D'Arcy, W. Janes, R. Gentz, H. J. Schoenfeld, C. Broger, H. Loetscher, and W. Lesslauer. 1993. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell 73:431-445. - PubMed
-
- Bogan, A. A., and K. S. Thorn. 1998. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280:1-9. - PubMed
-
- Browne, H., B. Bruun, and T. Minson. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J. Gen. Virol. 82:1419-1422. - PubMed
-
- Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

