Synergistic activation of human immunodeficiency virus type 1 promoter activity by NF-kappaB and inhibitors of deacetylases: potential perspectives for the development of therapeutic strategies
- PMID: 12368351
- PMCID: PMC136606
- DOI: 10.1128/jvi.76.21.11091-11103.2002
Synergistic activation of human immunodeficiency virus type 1 promoter activity by NF-kappaB and inhibitors of deacetylases: potential perspectives for the development of therapeutic strategies
Abstract
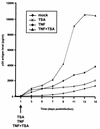
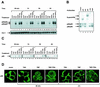
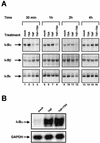
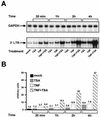
The transcription factor NF-kappaB plays a central role in the human immunodeficiency virus type 1 (HIV-1) activation pathway. HIV-1 transcription is also regulated by protein acetylation, since treatment with deacetylase inhibitors such as trichostatin A (TSA) or sodium butyrate (NaBut) markedly induces HIV-1 transcriptional activity of the long terminal repeat (LTR) promoter. Here, we demonstrate that TSA (NaBut) synergized with both ectopically expressed p50/p65 and tumor necrosis factor alpha/SF2 (TNF)-induced NF-kappaB to activate the LTR. This was confirmed for LTRs from subtypes A through G of the HIV-1 major group, with a positive correlation between the number of kappaB sites present in the LTRs and the amplitude of the TNF-TSA synergism. Mechanistically, TSA (NaBut) delayed the cytoplasmic recovery of the inhibitory protein IkappaBalpha. This coincided with a prolonged intranuclear presence and DNA binding activity of NF-kappaB. The physiological relevance of the TNF-TSA (NaBut) synergism was shown on HIV-1 replication in both acutely and latently HIV-infected cell lines. Therefore, our results open new therapeutic strategies aimed at decreasing or eliminating the pool of latently HIV-infected reservoirs by forcing viral expression.
Figures




References
-
- Adams, M., L. Sharmeen, J. Kimpton, J. M. Romeo, J. V. Garcia, B. M. Peterlin, M. Groudine, and M. Emerman. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. USA 91:3862-3866. - PMC - PubMed
-
- Chen, H., M. Tini, and R. M. Evans. 2001. HATs on and beyond chromatin. Curr. Opin. Cell Biol. 13:218-224. - PubMed
-
- Chen, L., W. Fischle, E. Verdin, and W. C. Greene. 2001. Duration of nuclear NF-κB action regulated by reversible acetylation. Science 293:1653-1657. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

