Comparative analysis of anti-hepatitis C virus activity and gene expression mediated by alpha, beta, and gamma interferons
- PMID: 12368359
- PMCID: PMC136623
- DOI: 10.1128/jvi.76.21.11148-11154.2002
Comparative analysis of anti-hepatitis C virus activity and gene expression mediated by alpha, beta, and gamma interferons
Abstract
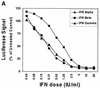
A direct comparison of the inhibitory effects of alpha, beta, and gamma interferons (IFNs) on replication of a hepatitis C virus subgenomic replicon in a hepatoma cell line revealed similarities in antiviral potency. However, alternate IFN-induced antiviral mechanisms were suggested following observations of striking differences between IFN-gamma and IFN-alpha/beta with respect to strength and durability of the antiviral response and the magnitude and pattern of IFN-mediated gene expression.
Figures


References
-
- Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. - PubMed
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
-
- Boehm, U., T. Klamp, M. Groot, and J. C. Howard. 1997. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 15:749-795. - PubMed
-
- Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

