Identification of a hepatitis B virus genome in wild chimpanzees (Pan troglodytes schweinfurthi) from East Africa indicates a wide geographical dispersion among equatorial African primates
- PMID: 12368360
- PMCID: PMC136620
- DOI: 10.1128/jvi.76.21.11155-11158.2002
Identification of a hepatitis B virus genome in wild chimpanzees (Pan troglodytes schweinfurthi) from East Africa indicates a wide geographical dispersion among equatorial African primates
Abstract
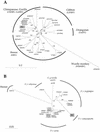
DNAs from four wild chimpanzees (Pan troglodytes schweinfurthi) from eastern Africa were screened for 14 DNA viruses and retroviruses. Between two and three viruses were found in each animal. An entire hepatitis B virus (HBV) genome was amplified and sequenced from samples taken from one animal. This indicates that HBV is distributed across the entire range of chimpanzee habitats.
Figures

References
-
- Bancroft, W. H., F. K. Mundon, and P. K. Russell. 1972. Detection of additional antigenic determinants of hepatitis B antigen. J. Immunol. 109:842-848. - PubMed
-
- Corbet, S., M. C. Müller-Trutwin, P. Versmisse, S. Delarue, A. Ayouba, J. Lewis, S. Brunak, P. Martin, F. Brun-Vezinet, F. Simon, F. Barre-Sinoussi, and P. Mauclere. 2000. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J. Virol. 74:529-534. - PMC - PubMed
-
- Courouce-Pauty, A. M., A. Plancon, and J. P. Soulier. 1983. Distribution of HBsAg subtypes in the world. Vox Sang. 44:197-211. - PubMed
-
- Greensill, J., J. A. Sheldon, K. K. Murthy, J. S. Bessonette, B. E. Beer, and T. F. Schulz. 2000. A chimpanzee rhadinovirus sequence related to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8: increased detection after HIV-1 infection in the absence of disease. AIDS 14:F129-F135. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical

