Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis
- PMID: 12368493
- PMCID: PMC151224
- DOI: 10.1105/tpc.004416
Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis
Abstract
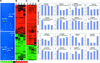
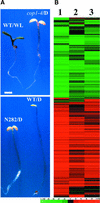
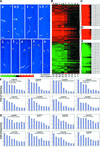
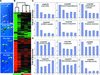
Microarray gene expression profiling was used to examine the role of COP1 in the light control of Arabidopsis genome expression. Qualitatively similar gene expression profiles were observed between wild-type seedlings grown in white light and multiple cop1 mutant alleles grown in the dark. Furthermore, overexpression of the dominant-negative-acting N terminus of COP1 (N282) in darkness produced a genome expression profile similar to those produced by white light and the cop1 mutations. Different cop1 mutant alleles, N282, and light treatment also resulted in distinct expression profiles in a small fraction of the genes examined. In the light, the genome expression of cop1 mutations displayed an exaggerated light response. COP1-regulated genes in the dark were estimated to account for >20% of the genome. Analysis of these COP1-regulated genes revealed that >28 cellular pathways are coordinately but antagonistically regulated by light and COP1. Interestingly, the gene expression regulation attributable to HY5 in the light is included largely within those genes regulated by COP1 in the dark. Thus, this genomic study supports the hypothesis that COP1 acts as a repressor of photomorphogenesis, possibly by controlling the degradation of transcription factors and their target gene expression. The majority of light-controlled genome expression could be accounted for by the negative regulation of COP1 activity.
Figures


Similar articles
-
The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis.Plant Cell. 2013 Apr;25(4):1243-57. doi: 10.1105/tpc.113.109751. Epub 2013 Apr 26. Plant Cell. 2013. PMID: 23624715 Free PMC article.
-
COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis.Plant Physiol. 2011 May;156(1):228-39. doi: 10.1104/pp.111.175042. Epub 2011 Mar 22. Plant Physiol. 2011. PMID: 21427283 Free PMC article.
-
Expression of an N-terminal fragment of COP1 confers a dominant-negative effect on light-regulated seedling development in Arabidopsis.Plant Cell. 1996 Sep;8(9):1491-503. doi: 10.1105/tpc.8.9.1491. Plant Cell. 1996. PMID: 8837504 Free PMC article.
-
The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling.Curr Opin Plant Biol. 2017 Jun;37:63-69. doi: 10.1016/j.pbi.2017.03.015. Epub 2017 Apr 21. Curr Opin Plant Biol. 2017. PMID: 28433946 Review.
-
The BBX subfamily IV: additional cogs and sprockets to fine-tune light-dependent development.Plant Signal Behav. 2013 Apr;8(4):e23831. doi: 10.4161/psb.23831. Epub 2013 Feb 20. Plant Signal Behav. 2013. PMID: 23425851 Free PMC article. Review.
Cited by
-
The peroxisome: orchestrating important developmental decisions from inside the cell.J Cell Biol. 2004 Mar 1;164(5):641-5. doi: 10.1083/jcb.200312081. Epub 2004 Feb 23. J Cell Biol. 2004. PMID: 14981090 Free PMC article. Review.
-
Plant biology research comes of age in China.Plant Cell. 2006 Nov;18(11):2855-64. doi: 10.1105/tpc.106.045393. Plant Cell. 2006. PMID: 17170389 Free PMC article. No abstract available.
-
Heterosis and polymorphisms of gene expression in an elite rice hybrid as revealed by a microarray analysis of 9198 unique ESTs.Plant Mol Biol. 2006 Nov;62(4-5):579-91. doi: 10.1007/s11103-006-9040-z. Epub 2006 Aug 29. Plant Mol Biol. 2006. PMID: 16941221
-
An initial biochemical and cell biological characterization of the mammalian homologue of a central plant developmental switch, COP1.BMC Cell Biol. 2002 Dec 4;3:30. doi: 10.1186/1471-2121-3-30. Epub 2002 Dec 4. BMC Cell Biol. 2002. PMID: 12466024 Free PMC article.
-
The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses.Plant Cell. 2003 May;15(5):1083-94. doi: 10.1105/tpc.010207. Plant Cell. 2003. PMID: 12724535 Free PMC article.
References
-
- Ang, L.H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. - PubMed
-
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

