The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat
- PMID: 12368498
- PMCID: PMC151229
- DOI: 10.1105/tpc.004127
The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat
Abstract
Screening for seed pigmentation phenotypes in Arabidopsis led to the isolation of three allelic yellow-seeded mutants, which defined the novel TRANSPARENT TESTA16 (TT16) locus. Cloning of TT16 was performed by T-DNA tagging and confirmed by genetic complementation and sequencing of two mutant alleles. TT16 encodes the ARABIDOPSIS BSISTER (ABS) MADS domain protein. ABS belongs to the recently identified "B-sister" (B(S)) clade, which contains genes of unknown function that are expressed mainly in female organs. Phylogenetic analyses using a maximum parsimony approach confirmed that TT16/ABS and related proteins form a monophyletic group. TT16/ABS was expressed mainly in the ovule, as are the other members of the B(S) clade. TT16/ABS is necessary for BANYULS expression and proanthocyanidin accumulation in the endothelium of the seed coat, with the exception of the chalazal-micropylar area. In addition, mutant phenotype and ectopic expression analyses suggested that TT16/ABS also is involved in the specification of endothelial cells. Nevertheless, TT16/ABS apparently is not required for proper ovule function. We report the functional characterization of a member of the B(S) MADS box gene subfamily, demonstrating its involvement in endothelial cell specification as well as in the increasingly complex genetic control of flavonoid biosynthesis in the Arabidopsis seed coat.
Figures
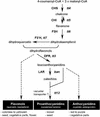
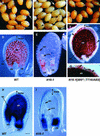



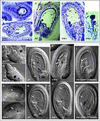

References
-
- Albert, S., Delseny, M., and Devic, M. (1997). BANYULS, a novel negative regulator of flavonoid biosynthesis in the Arabidopsis seed coat. Plant J. 11, 289–299. - PubMed
-
- Alvarez-Buylla, E.R., Pelaz, S., Liljegren, S.J., Gold, S.E., Burgeff, C., Ditta, G.S., Ribas de Pouplana, L., Martinez-Castilla, L., and Yanofsky, M.F. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97, 5328–5333. - PMC - PubMed
-
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199.
-
- Becker, A., Kaufmann, K., Freialdenhoven, A., Vincent, C., Li, M.-A., Saedler, H., and Theissen, G. (2002). A novel MADS-box gene subfamily with a sister-group relationship to class B floral homeotic genes. Mol. Genet. Genomics 266, 942–950. - PubMed
-
- Becker, A., Winter, K.-U., Meyer, B., Saedler, H., and Theissen, G. (2000). MADS-box gene diversity in seed plants 300 million years ago. Mol. Biol. Evol. 17, 1425–1434. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

