High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1
- PMID: 12368501
- PMCID: PMC151232
- DOI: 10.1105/tpc.004218
High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1
Abstract
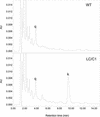
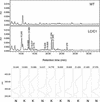
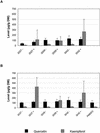
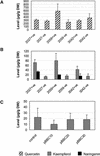
Flavonoids are a group of polyphenolic plant secondary metabolites important for plant biology and human nutrition. In particular flavonols are potent antioxidants, and their dietary intake is correlated with a reduced risk of cardiovascular diseases. Tomato fruit contain only in their peel small amounts of flavonoids, mainly naringenin chalcone and the flavonol rutin, a quercetin glycoside. To increase flavonoid levels in tomato, we expressed the maize transcription factor genes LC and C1 in the fruit of genetically modified tomato plants. Expression of both genes was required and sufficient to upregulate the flavonoid pathway in tomato fruit flesh, a tissue that normally does not produce any flavonoids. These fruit accumulated high levels of the flavonol kaempferol and, to a lesser extent, the flavanone naringenin in their flesh. All flavonoids detected were present as glycosides. Anthocyanins, previously reported to accumulate upon LC expression in several plant species, were present in LC/C1 tomato leaves but could not be detected in ripe LC/C1 fruit. RNA expression analysis of ripening fruit revealed that, with the exception of chalcone isomerase, all of the structural genes required for the production of kaempferol-type flavonols and pelargonidin-type anthocyanins were induced strongly by the LC/C1 transcription factors. Expression of the genes encoding flavanone-3'-hydroxylase and flavanone-3'5'-hydroxylase, which are required for the modification of B-ring hydroxylation patterns, was not affected by LC/C1. Comparison of flavonoid profiles and gene expression data between tomato leaves and fruit indicates that the absence of anthocyanins in LC/C1 fruit is attributable primarily to an insufficient expression of the gene encoding flavanone-3'5'-hydroxylase, in combination with a strong preference of the tomato dihydroflavonol reductase enzyme to use the flavanone-3'5'-hydroxylase reaction product dihydromyricetin as a substrate.
Figures







References
-
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20, 1195–1197. - PubMed
-
- Bongue-Bartelsman, M., and Philips, D.A. (1995). Nitrogen stress regulates gene expression of enzymes in the flavonoid biosynthetic pathway of tomato. Plant Physiol. Biochem. 33, 539–546.
-
- Bovy, A., van den Berg, C., de Vrieze, G., Thompson, W.F., Weisbeek, P., and Smeekens, S. (1995). Light-regulated expression of the Arabidopsis thaliana ferredoxin gene requires sequences upstream and downstream of the transcription initiation site. Plant Mol. Biol. 27, 27–39. - PubMed
-
- Bovy, A.G., Hijden, H.T., Hughes, S.G., Muir, S.R., Tunen, A.J., Verhoezen, M.E., and Vos, C.H. (1999). Methods and composition for modulating flavonoid content. W09937794 A 19990729.
-
- Bradley, J.M., Davies, K.M., Deroles, S.C., Bloor, S.J., and Lewis, D.H. (1998). The maize Lc regulatory gene up-regulates the flavonoid biosynthetic pathway of petunia. Plant J. 13, 381–392.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

