A polyamine metabolon involving aminopropyl transferase complexes in Arabidopsis
- PMID: 12368503
- PMCID: PMC151234
- DOI: 10.1105/tpc.004077
A polyamine metabolon involving aminopropyl transferase complexes in Arabidopsis
Abstract
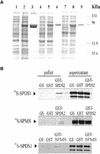
The conversion of putrescine to spermidine in the biosynthetic pathway of plant polyamines is catalyzed by two closely related spermidine synthases, SPDS1 and SPDS2, in Arabidopsis. In the yeast two-hybrid system, SPDS2 was found to interact with SPDS1 and a novel protein, SPMS (spermine synthase), which is homologous with SPDS2 and SPDS1. SPMS interacts with both SPDS1 and SPDS2 in yeast and in vitro. Unlike SPDS1 and SPDS2, SPMS failed to suppress the speDelta3 deficiency of spermidine synthase in yeast. However, SPMS was able to complement the speDelta4 spermine deficiency in yeast, indicating that SPMS is a novel spermine synthase. The SPDS and SPMS proteins showed no homodimerization but formed heterodimers in vitro. Pairwise coexpression of hemagglutinin- and c-Myc epitope-labeled proteins in Arabidopsis cells confirmed the existence of coimmunoprecipitating SPDS1-SPDS2 and SDPS2-SPMS heterodimers in vivo. The epitope-labeled SPDS and SPMS proteins copurified with protein complexes ranging in size from 650 to 750 kD. Our data demonstrate the existence of a metabolon involving at least the last two steps of polyamine biosynthesis in Arabidopsis.
Figures




Similar articles
-
Characterization of the spermidine synthase-related gene family in Arabidopsis thaliana.FEBS Lett. 2002 Sep 11;527(1-3):176-80. doi: 10.1016/s0014-5793(02)03217-9. FEBS Lett. 2002. PMID: 12220656
-
Spermidine synthase genes are essential for survival of Arabidopsis.Plant Physiol. 2004 Jul;135(3):1565-73. doi: 10.1104/pp.104.041699. Epub 2004 Jul 9. Plant Physiol. 2004. PMID: 15247389 Free PMC article.
-
Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells.PLoS One. 2012;7(10):e46907. doi: 10.1371/journal.pone.0046907. Epub 2012 Oct 8. PLoS One. 2012. PMID: 23056524 Free PMC article.
-
Polyamines.Annu Rev Biochem. 1984;53:749-90. doi: 10.1146/annurev.bi.53.070184.003533. Annu Rev Biochem. 1984. PMID: 6206782 Review. No abstract available.
-
Modulation of learning and memory by natural polyamines.Pharmacol Res. 2016 Oct;112:99-118. doi: 10.1016/j.phrs.2016.03.023. Epub 2016 Mar 22. Pharmacol Res. 2016. PMID: 27015893 Review.
Cited by
-
Arabidopsis spermidine synthase is targeted by an effector protein of the cyst nematode Heterodera schachtii.Plant Physiol. 2010 Feb;152(2):968-84. doi: 10.1104/pp.109.150557. Epub 2009 Dec 4. Plant Physiol. 2010. PMID: 19965964 Free PMC article.
-
Spermine synthase.Cell Mol Life Sci. 2010 Jan;67(1):113-21. doi: 10.1007/s00018-009-0165-5. Epub 2009 Oct 27. Cell Mol Life Sci. 2010. PMID: 19859664 Free PMC article. Review.
-
Zmspds2 maize gene: Coding a spermine synthase?Plant Signal Behav. 2008 Aug;3(8):551-3. doi: 10.4161/psb.3.8.5697. Plant Signal Behav. 2008. PMID: 19704464 Free PMC article.
-
The Spermine Synthase OsSPMS1 Regulates Seed Germination, Grain Size, and Yield.Plant Physiol. 2018 Dec;178(4):1522-1536. doi: 10.1104/pp.18.00877. Epub 2018 Sep 6. Plant Physiol. 2018. PMID: 30190417 Free PMC article.
-
Polyamines: molecules with regulatory functions in plant abiotic stress tolerance.Planta. 2010 May;231(6):1237-49. doi: 10.1007/s00425-010-1130-0. Epub 2010 Mar 11. Planta. 2010. PMID: 20221631 Review.
References
-
- Alabadí, D., and Carbonell, J. (1999). Differential expression of two spermidine synthase genes during early fruit development and in vegetative tissues of pea. Plant Mol. Biol. 39, 933–943. - PubMed
-
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1989). Current Protocols in Molecular Biology. (New York: Greene/Wiley).
-
- Bowman, W.H., Tabor, C.W., and Tabor, H. (1973). Spermidine biosynthesis: Purification and properties of propylamine transferase from Escherichia coli. J. Biol. Chem. 248, 2480–2486. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

