MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function
- PMID: 12370088
- PMCID: PMC129983
- DOI: 10.1186/1471-2091-3-28
MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function
Abstract
Background: Three macromolecular assemblages, the lid complex of the proteasome, the COP9-Signalosome (CSN) and the eIF3 complex, all consist of multiple proteins harboring MPN and PCI domains. Up to now, no specific function for any of these proteins has been defined, nor has the importance of these motifs been elucidated. In particular Rpn11, a lid subunit, serves as the paradigm for MPN-containing proteins as it is highly conserved and important for proteasome function.
Results: We have identified a sequence motif, termed the MPN+ motif, which is highly conserved in a subset of MPN domain proteins such as Rpn11 and Csn5/Jab1, but is not present outside of this subfamily. The MPN+ motif consists of five polar residues that resemble the active site residues of hydrolytic enzyme classes, particularly that of metalloproteases. By using site-directed mutagenesis, we show that the MPN+ residues are important for the function of Rpn11, while a highly conserved Cys residue outside of the MPN+ motif is not essential. Single amino acid substitutions in MPN+ residues all show similar phenotypes, including slow growth, sensitivity to temperature and amino acid analogs, and general proteasome-dependent proteolysis defects.
Conclusions: The MPN+ motif is abundant in certain MPN-domain proteins, including newly identified proteins of eukaryotes, bacteria and archaea thought to act outside of the traditional large PCI/MPN complexes. The putative catalytic nature of the MPN+ motif makes it a good candidate for a pivotal enzymatic function, possibly a proteasome-associated deubiquitinating activity and a CSN-associated Nedd8/Rub1-removing activity.
Figures


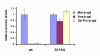

References
-
- Glickman MH, Ciechanover A. The Ubiquitin-proteasome Proteolytic Pathway: Destruction for the sake of construction. Physiol Rev. 2002;82:373–428. - PubMed
-
- D'Andrea A, Pellman D. Deubiquitinating enzymes: A new class of biological regulators. Crit Rev Biochem Mol Biol. 1998;33:337–332. - PubMed
-
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9/Signalosome and eIF3. Cell. 1998;94:615–623. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

