The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots
- PMID: 12370409
- PMCID: PMC129801
- DOI: 10.1073/pnas.212448699
The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots
Abstract
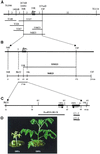
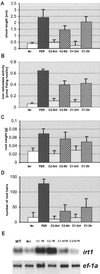
Iron deficiency is among the most common nutritional disorders in plants. To cope with low iron supply, plants with the exception of the Gramineae increase the solubility and uptake of iron by inducing physiological and developmental alterations including iron reduction, soil acidification, Fe(II) transport and root-hair proliferation (strategy I). The chlorotic tomato fer mutant fails to activate the strategy I. It was shown previously that the fer gene is required in the root. Here, we show that fer plants exhibit root developmental phenotypes after low and sufficient iron nutrition indicating that FER acts irrespective of iron supply. Mutant fer roots displayed lower Leirt1 expression than wild-type roots. We isolated the fer gene by map-based cloning and demonstrate that it encodes a protein containing a basic helix-loop-helix domain. fer is expressed in a cell-specific pattern at the root tip independently from iron supply. Our results suggest that FER may control root physiology and development at a transcriptional level in response to iron supply and thus may be the first identified regulator for iron nutrition in plants.
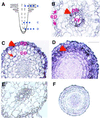
Figures
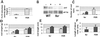

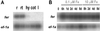
References
-
- Guerinot M L. Science. 2000;287:241–243. - PubMed
-
- World Health Organization. Battling Iron Deficiency Anaemia. 2002. www.who.int/nut/ida.htm , www.who.int/nut/ida.htm. .
-
- Wu J, Boyle E, Sunda W, Wen L-S. Science. 2001;293:847–849. - PubMed
-
- Geider R J. Nature. 1999;400:815–816.
-
- Guerinot M L. Nat Biotechnol. 2001;19:417–418. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

