Genomic analysis of membrane protein families: abundance and conserved motifs
- PMID: 12372142
- PMCID: PMC134483
- DOI: 10.1186/gb-2002-3-10-research0054
Genomic analysis of membrane protein families: abundance and conserved motifs
Abstract
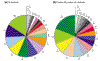
Background: Polytopic membrane proteins can be related to each other on the basis of the number of transmembrane helices and sequence similarities. Building on the Pfam classification of protein domain families, and using transmembrane-helix prediction and sequence-similarity searching, we identified a total of 526 well-characterized membrane protein families in 26 recently sequenced genomes. To this we added a clustering of a number of predicted but unclassified membrane proteins, resulting in a total of 637 membrane protein families.
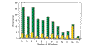
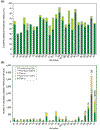
Results: Analysis of the occurrence and composition of these families revealed several interesting trends. The number of assigned membrane protein domains has an approximately linear relationship to the total number of open reading frames (ORFs) in 26 genomes studied. Caenorhabditis elegans is an apparent outlier, because of its high representation of seven-span transmembrane (7-TM) chemoreceptor families. In all genomes, including that of C. elegans, the number of distinct membrane protein families has a logarithmic relation to the number of ORFs. Glycine, proline, and tyrosine locations tend to be conserved in transmembrane regions within families, whereas isoleucine, valine, and methionine locations are relatively mutable. Analysis of motifs in putative transmembrane helices reveals that GxxxG and GxxxxxxG (which can be written GG4 and GG7, respectively; see Materials and methods) are among the most prevalent. This was noted in earlier studies; we now find these motifs are particularly well conserved in families, however, especially those corresponding to transporters, symporters, and channels.
Conclusions: We carried out a genome-wide analysis on patterns of the classified polytopic membrane protein families and analyzed the distribution of conserved amino acids and motifs in the transmembrane helix regions in these families.
Figures


References
-
- Paulsen IT, Sliwinski MK, Saier MHJ. Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J Mol Biol. 1998;277:573–592. - PubMed
-
- Paulsen IT, Nguyen L, Sliwinski MK, Rabus R, Saier MHJ. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J Mol Biol. 2000;301:75–100. - PubMed
-
- Gerstein M. A structural census of genomes: comparing bacterial, eukaryotic, and archaeal genomes in terms of protein structure. J Mol Biol. 1997;274:562–576. - PubMed
-
- Gerstein M. Patterns of protein-fold usage in eight microbial genomes: a comprehensive structural census. Proteins. 1998;33:518–534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

