Asymmetric directional mutation pressures in bacteria
- PMID: 12372146
- PMCID: PMC134625
- DOI: 10.1186/gb-2002-3-10-research0058
Asymmetric directional mutation pressures in bacteria
Abstract
Background: When there are no strand-specific biases in mutation and selection rates (that is, in the substitution rates) between the two strands of DNA, the average nucleotide composition is theoretically expected to be A = T and G = C within each strand. Deviations from these equalities are therefore evidence for an asymmetry in selection and/or mutation between the two strands. By focusing on weakly selected regions that could be oriented with respect to replication in 43 out of 51 completely sequenced bacterial chromosomes, we have been able to detect asymmetric directional mutation pressures.
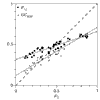
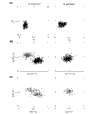
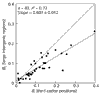
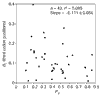
Results: Most of the 43 chromosomes were found to be relatively enriched in G over C and T over A, and slightly depleted in G+C, in their weakly selected positions (intergenic regions and third codon positions) in the leading strand compared with the lagging strand. Deviations from A = T and G = C were highly correlated between third codon positions and intergenic regions, with a lower degree of deviation in intergenic regions, and were not correlated with overall genomic G+C content.
Conclusions: During the course of bacterial chromosome evolution, the effects of asymmetric directional mutation pressures are commonly observed in weakly selected positions. The degree of deviation from equality is highly variable among species, and within species is higher in third codon positions than in intergenic regions. The orientation of these effects is almost universal and is compatible in most cases with the hypothesis of an excess of cytosine deamination in the single-stranded state during DNA replication. However, the variation in G+C content between species is influenced by factors other than asymmetric mutation pressure.
Figures

Similar articles
-
Wide intra-genomic G+C heterogeneity in human and chicken is mainly due to strand-symmetric directional mutation pressures: dGTP-oxidation and symmetric cytosine-deamination hypotheses.Gene. 2002 Oct 30;300(1-2):141-54. doi: 10.1016/s0378-1119(02)01046-6. Gene. 2002. PMID: 12468095
-
Atypical at skew in Firmicute genomes results from selection and not from mutation.PLoS Genet. 2011 Sep;7(9):e1002283. doi: 10.1371/journal.pgen.1002283. Epub 2011 Sep 15. PLoS Genet. 2011. PMID: 21935355 Free PMC article.
-
Asymmetric substitution patterns in the two DNA strands of bacteria.Mol Biol Evol. 1996 May;13(5):660-5. doi: 10.1093/oxfordjournals.molbev.a025626. Mol Biol Evol. 1996. PMID: 8676740
-
Polarisation of prokaryotic chromosomes.Curr Opin Microbiol. 2003 Apr;6(2):101-8. doi: 10.1016/s1369-5274(03)00024-9. Curr Opin Microbiol. 2003. PMID: 12732297 Review.
-
Random versus Cell Cycle-Regulated Replication Initiation in Bacteria: Insights from Studying Vibrio cholerae Chromosome 2.Microbiol Mol Biol Rev. 2016 Nov 30;81(1):e00033-16. doi: 10.1128/MMBR.00033-16. Print 2017 Mar. Microbiol Mol Biol Rev. 2016. PMID: 27903655 Free PMC article. Review.
Cited by
-
Two common profiles exist for genomic oligonucleotide frequencies.BMC Res Notes. 2012 Nov 17;5:639. doi: 10.1186/1756-0500-5-639. BMC Res Notes. 2012. PMID: 23158698 Free PMC article.
-
Conflict between translation initiation and elongation in vertebrate mitochondrial genomes.PLoS One. 2007 Feb 21;2(2):e227. doi: 10.1371/journal.pone.0000227. PLoS One. 2007. PMID: 17311091 Free PMC article.
-
Modeling compositional dynamics based on GC and purine contents of protein-coding sequences.Biol Direct. 2010 Nov 8;5:63. doi: 10.1186/1745-6150-5-63. Biol Direct. 2010. PMID: 21059261 Free PMC article.
-
Codon usage analysis of photolyase encoding genes of cyanobacteria inhabiting diverse habitats.3 Biotech. 2017 Jul;7(3):192. doi: 10.1007/s13205-017-0826-2. Epub 2017 Jun 29. 3 Biotech. 2017. PMID: 28664377 Free PMC article.
-
Asymmetric Context-Dependent Mutation Patterns Revealed through Mutation-Accumulation Experiments.Mol Biol Evol. 2015 Jul;32(7):1672-83. doi: 10.1093/molbev/msv055. Epub 2015 Mar 6. Mol Biol Evol. 2015. PMID: 25750180 Free PMC article.
References
-
- Lee KY, Wahl R, Barbu E. Contenu en bases puriques et pyrimidiques des acides désoxyribonucléiques des bactéries. Ann Inst Pasteur. 1956;91:212–224. - PubMed
-
- Belozersky AN, Spirin AS. A correlation between the compositions of deoxyribonucleic and ribonucleic acids. Nature. 1958;182:111–112. - PubMed
-
- Sueoka N, Marmur J, Doty P. Heterogeneity in deoxyriboneucleic acids. II. Dependence of the density of deoxyribonucleic acids on guanine-cytosine. Nature. 1959;183:1427–1431. - PubMed
-
- Kerr ARW, Peden JF, Sharp PM. Systematic base composition variation around the genome of Mycoplasma genitalium, but not Mycoplasma pneumoniae. Mol Microbiol. 1997;25:1177–1179. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

