Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase
- PMID: 12372305
- PMCID: PMC2713760
- DOI: 10.1016/s0092-8674(02)00999-6
Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase
Abstract
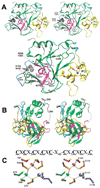
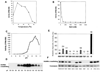
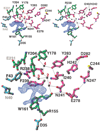
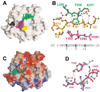
AdoMet-dependent methylation of histones is part of the "histone code" that can profoundly influence gene expression. We describe the crystal structure of Neurospora DIM-5, a histone H3 lysine 9 methyltranferase (HKMT), determined at 1.98 A resolution, as well as results of biochemical characterization and site-directed mutagenesis of key residues. This SET domain protein bears no structural similarity to previously characterized AdoMet-dependent methyltransferases but includes notable features such as a triangular Zn3Cys9 zinc cluster in the pre-SET domain and a AdoMet binding site in the SET domain essential for methyl transfer. The structure suggests a mechanism for the methylation reaction and provides the structural basis for functional characterization of the HKMT family and the SET domain.
Figures


References
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
-
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. - PMC - PubMed
-
- Blumenthal RM, Cheng X. A Taq attack displaces bases. Nat. Struct. Biol. 2001;8:101–103. - PubMed
-
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 2002;30:73–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

