Genomic deletions of MSH2 and MLH1 in colorectal cancer families detected by a novel mutation detection approach
- PMID: 12373605
- PMCID: PMC2376172
- DOI: 10.1038/sj.bjc.6600565
Genomic deletions of MSH2 and MLH1 in colorectal cancer families detected by a novel mutation detection approach
Abstract
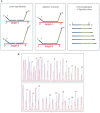
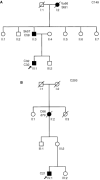
Hereditary non-polyposis colorectal cancer is an autosomal dominant condition due to germline mutations in DNA-mismatch-repair genes, in particular MLH1, MSH2 and MSH6. Here we describe the application of a novel technique for the detection of genomic deletions in MLH1 and MSH2. This method, called multiplex ligation-dependent probe amplification, is a quantitative multiplex PCR approach to determine the relative copy number of each MLH1 and MSH2 exon. Mutation screening of genes was performed in 126 colorectal cancer families selected on the basis of clinical criteria and in addition, for a subset of families, the presence of microsatellite instability (MSI-high) in tumours. Thirty-eight germline mutations were detected in 37 (29.4%) of these kindreds, 31 of which have a predicted pathogenic effect. Among families with MSI-high tumours 65.7% harboured germline gene defects. Genomic deletions accounted for 54.8% of the pathogenic mutations. A complete deletion of the MLH1 gene was detected in two families. The multiplex ligation-dependent probe amplification approach is a rapid method for the detection of genomic deletions in MLH1 and MSH2. In addition, it reveals alterations that might escape detection using conventional diagnostic techniques. Multiplex ligation-dependent probe amplification might be considered as an early step in the molecular diagnosis of hereditary non-polyposis colorectal cancer.
Copyright 2002 Cancer Research UK
Figures
References
-
- BapatBVMadlenskyLTempleLKFHirukiTRedstonMBaronDLXiaLMarcusVASoraviaCMitriAShenWGryfeRBerkTChodirkerBNCohenZGallingerS1999Family history characteristics, tumor microsatellite instability and germline MSH2 and MLH1 mutations in hereditary colorectal cancer Hum Genet 104167176 - PubMed
-
- BolandCRThibodeauSNHamiltonSRSidranskyDEshlemanJRBurtRWMeltzerSJRodriguez-BigazMAFoddeRRanzaniGNSrivastavaS1998A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer Cancer Res 5852485257 - PubMed
-
- CharbonnierFRauxGWangQDrouotNCordierFLimacherJ-MSaurinJ-CPuisieuxAOlschwangSFrebourgT2000Detection of exon deletions and duplications of the mismatch repair genes in hereditary nonpolyposis colorectal cancer families using multiplex polymerase chain reaction of short fluorescent fragments Cancer Res 6027602763 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

