60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm
- PMID: 12374754
- PMCID: PMC129079
- DOI: 10.1093/emboj/cdf547
60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm
Abstract
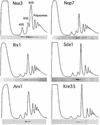
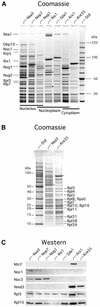
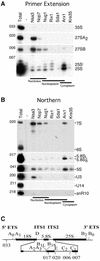
60S ribosomes undergo initial assembly in the nucleolus before export to the cytoplasm and recent analyses have identified several nucleolar pre-60S particles. To unravel the steps in the pathway of ribosome formation, we have purified the pre-60S ribosomes associated with proteins predicted to act at different stages as the pre-ribosomes transit from the nucleolus through the nucleoplasm and are then exported to the cytoplasm for final maturation. About 50 non-ribosomal proteins are associated with the early nucleolar pre-60S ribosomes. During subsequent maturation and transport to the nucleoplasm, many of these factors are removed, while others remain attached and additional factors transiently associate. When the 60S precursor particles are close to exit from the nucleus they associate with at least two export factors, Nmd3 and Mtr2. As the 60S pre-ribosome reaches the cytoplasm, almost all of the factors are dissociated. These data provide an initial biochemical map of 60S ribosomal subunit formation on its path from the nucleolus to the cytoplasm.
Figures


References
-
- Baßler J., Grandi,P., Gadal,O., Leßmann,T., Tollervey,D., Lechner,J. and Hurt,E.C. (2001) Identification of a 60S pre-ribosomal particle that is closely linked to nuclear export. Mol. Cell, 8, 517–529. - PubMed
-
- Buscemi G., Saracino,F., Masnada,D. and Carbone,M.L. (2000) The Saccharomyces cerevisiae SDA1 gene is required for actin cytoskeleton organization and cell cycle progression. J. Cell Sci., 113, 1199–1211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

