Site-specific ORC binding, pre-replication complex assembly and DNA synthesis at Schizosaccharomyces pombe replication origins
- PMID: 12374757
- PMCID: PMC129078
- DOI: 10.1093/emboj/cdf546
Site-specific ORC binding, pre-replication complex assembly and DNA synthesis at Schizosaccharomyces pombe replication origins
Abstract
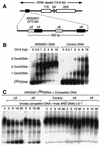
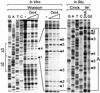
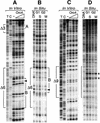
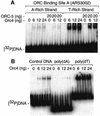
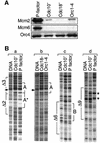
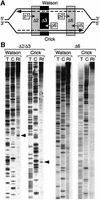
Previous studies have shown that the Schizo saccharomyces pombe Orc4 subunit is solely responsible for in vitro binding of origin recognition complex (ORC) to specific AT-rich sites within S.pombe replication origins. Using ARS3001, a S.pombe replication origin consisting of four genetically required sites, we show that, in situ as well as in vitro, Orc4 binds strongly to the Delta3 site, weakly to the Delta6 site and not at all to the remaining sequences. In situ, the footprint over Delta3 is extended during G(1) phase, but only when Cdc18 is present and Mcm proteins are bound to chromatin. Moreover, this footprint extends into the adjacent Delta2 site, where leading strand DNA synthesis begins. Therefore, we conclude that ARS3001 consists of a single primary ORC binding site that assembles a pre-replication complex and initiates DNA synthesis, plus an additional novel origin element (Delta9) that neither binds ORC nor functions as a centromere, but does bind an as yet unidentified protein throughout the cell cycle. Schizosaccharomyces pombe may be an appropriate paradigm for the complex origins found in the metazoa.
Figures


Similar articles
-
Sap1 is a replication-initiation factor essential for the assembly of pre-replicative complex in the fission yeast Schizosaccharomyces pombe.J Biol Chem. 2017 Apr 14;292(15):6056-6075. doi: 10.1074/jbc.M116.767806. Epub 2017 Feb 21. J Biol Chem. 2017. PMID: 28223353 Free PMC article.
-
Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit.Mol Cell Biol. 2001 Dec;21(23):8095-103. doi: 10.1128/MCB.21.23.8095-8103.2001. Mol Cell Biol. 2001. PMID: 11689699 Free PMC article.
-
Xenopus origin recognition complex (ORC) initiates DNA replication preferentially at sequences targeted by Schizosaccharomyces pombe ORC.EMBO J. 2003 Jul 1;22(13):3441-50. doi: 10.1093/emboj/cdg319. EMBO J. 2003. PMID: 12840006 Free PMC article.
-
Stepwise assembly of initiation complexes at budding yeast replication origins during the cell cycle.J Cell Sci Suppl. 1995;19:67-72. doi: 10.1242/jcs.1995.supplement_19.9. J Cell Sci Suppl. 1995. PMID: 8655649 Review.
-
Specification of DNA replication origins and genomic base composition in fission yeasts.J Mol Biol. 2013 Nov 29;425(23):4706-13. doi: 10.1016/j.jmb.2013.09.023. Epub 2013 Oct 3. J Mol Biol. 2013. PMID: 24095860 Review.
Cited by
-
Once in a lifetime: strategies for preventing re-replication in prokaryotic and eukaryotic cells.EMBO Rep. 2008 Feb;9(2):151-6. doi: 10.1038/sj.embor.2008.2. EMBO Rep. 2008. PMID: 18246107 Free PMC article. Review.
-
The chromatin backdrop of DNA replication: lessons from genetics and genome-scale analyses.Biochim Biophys Acta. 2012 Jul;1819(7):794-801. doi: 10.1016/j.bbagrm.2012.01.017. Epub 2012 Feb 8. Biochim Biophys Acta. 2012. PMID: 22342530 Free PMC article. Review.
-
DNA elements modulating the KARS12 chromosomal replicator in Kluyveromyces lactis.Mol Genet Genomics. 2007 Mar;277(3):287-99. doi: 10.1007/s00438-006-0188-7. Epub 2006 Nov 29. Mol Genet Genomics. 2007. PMID: 17136349
-
A novel intermediate in initiation complex assembly for fission yeast DNA replication.Mol Biol Cell. 2004 Aug;15(8):3740-50. doi: 10.1091/mbc.e04-04-0292. Epub 2004 Jun 11. Mol Biol Cell. 2004. PMID: 15194812 Free PMC article.
-
Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe.EMBO Rep. 2003 Nov;4(11):1048-53. doi: 10.1038/sj.embor.embor7400008. Epub 2003 Oct 17. EMBO Rep. 2003. PMID: 14566325 Free PMC article.
References
-
- Abdurashidova G., Deganuto,M., Klima,R., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science, 287, 2023–2026. - PubMed
-
- Aladjem M.I., Rodewald,L.W., Kolman,J.L. and Wahl,G.M. (1998) Genetic dissection of a mammalian replicator in the human β-globin locus. Science, 281, 1005–1009. - PubMed
-
- Aladjem M.I., Rodewald,L.W., Lin,C.M., Bowman,S., Cimbora,D.M., Brody,L.L., Epner,E.M., Groudine,M. and Wahl,G.M. (2002) Replication initiation patterns in the β-globin loci of totipotent and differentiated murine cells: evidence for multiple initiation regions. Mol. Cell. Biol., 22, 442–452. - PMC - PubMed
-
- Bell S.P. (2002) The origin recognition complex: from simple origins to complex functions. Genes Dev., 16, 659–672. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

