Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence
- PMID: 12374814
- PMCID: PMC135393
- DOI: 10.1128/JB.184.21.5826-5832.2002
Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence
Abstract
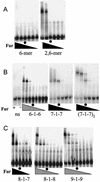
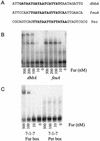
Ferric uptake repressor (Fur) proteins regulate the expression of iron homeostasis genes in response to intracellular iron levels. In general, Fur proteins bind with high affinity to a 19-bp inverted repeat sequence known as the Fur box. An alignment of 19 operator sites recognized by Bacillus subtilis Fur revealed a different conserved 15-bp (7-1-7) inverted repeat present twice within this 19-bp consensus sequence. We demonstrated using electrophoretic mobility shift assays that this 7-1-7 inverted repeat comprises a minimal recognition site for high-affinity binding by Fur. The resulting revised consensus sequence is remarkably similar to a related 7-1-7 inverted repeat sequence recognized by PerR, a Fur paralog. Our analysis of the affinity and stoichiometry of DNA binding by B. subtilis Fur, together with a reinterpretation of previously described studies of Escherichia coli Fur, supports a model in which the 19-bp Fur box represents overlapping recognition sites for two Fur dimers bound to opposite faces of the DNA helix. The resulting recognition complex is reminiscent of that observed for the functionally related protein DtxR. Like Fur, DtxR contains a helix-turn-helix DNA-binding motif, recognizes a 19-bp inverted repeat sequence, and has a typical DNase I footprint of approximately 30 bp. By envisioning a similar mode of DNA recognition for Fur, we can account for the internal symmetries noted previously within the Fur box, the tendency of Fur to extend into adjacent regions of DNA in a sequence-selective manner, and the observed patterns of DNA protection against enzymatic and chemical probes.
Figures



References
-
- Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol., in press. - PubMed
-
- Bell, S. D., S. S. Cairns, R. L. Robson, and S. P. Jackson. 1999. Transcriptional regulation of an archaeal operon in vivo and in vitro. Mol. Cell 4:971-982. - PubMed
-
- Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

