SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria
- PMID: 12374852
- PMCID: PMC129731
- DOI: 10.1073/pnas.222538099
SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria
Abstract
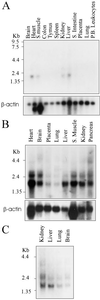
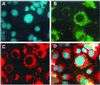
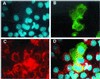
The SIR2 (silent information regulator 2) gene family has diverse functions in yeast including gene silencing, DNA repair, cell-cycle progression, and chromosome fidelity in meiosis and aging. Human homologues, termed sirtuins, are highly conserved but are of unknown function. We previously identified a large imprinted gene domain on 11p15.5 and investigated the 11p15.5 sirtuin SIRT3. Although this gene was not imprinted, we found that it is localized to mitochondria, with a mitochondrial targeting signal within a unique N-terminal peptide sequence. The encoded protein was found also to possess NAD(+)-dependent histone deacetylase activity. These results suggest a previously unrecognized organelle for sirtuin function and that the role of SIRT3 in mitochondria involves protein deacetylation.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

